7Z7E
 
 | | Crystal structure of p63 DNA binding domain in complex with inhibitory DARPin G4 | | 分子名称: | DARPIN, Isoform 4 of Tumor protein 63, ZINC ION | | 著者 | Strubel, A, Gebel, J, Chaikuad, A, Muenick, P, Doetsch, V. | | 登録日 | 2022-03-15 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
5ICG
 
 | | Crystal structure of apo (S)-norcoclaurine 6-O-methyltransferase | | 分子名称: | (S)-norcoclaurine 6-O-methyltransferase, POTASSIUM ION | | 著者 | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | 登録日 | 2016-02-23 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
8OI4
 
 | | Metagenomic Beta-galactosidase from Glycoside Hydrolase family GH154 | | 分子名称: | Beta-galactosidase, CHLORIDE ION, GLYCEROL | | 著者 | Pijning, T, Hameleers, L, Jurak, E, Guskov, A. | | 登録日 | 2023-03-22 | | 公開日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Novel beta-galactosidase activity and first crystal structure of Glycoside Hydrolase family 154.
N Biotechnol, 80, 2023
|
|
5A2B
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | 分子名称: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | 登録日 | 2015-05-17 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5AQA
 
 | | DARPin-based Crystallization Chaperones exploit Molecular Geometry as a Screening Dimension in Protein Crystallography | | 分子名称: | OFF7_DB04V3, THIOCYANATE ION | | 著者 | Batyuk, A, Wu, Y, Honegger, A, Heberling, M, Plueckthun, A. | | 登録日 | 2015-09-21 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Darpin-Based Crystallization Chaperones Exploit Molecular Geometry as a Screening Dimension in Protein Crystallography
J.Mol.Biol., 428, 2016
|
|
7Z21
 
 | | BAF A12T bound to the lamin A/C Ig-fold domain | | 分子名称: | Barrier-to-autointegration factor, N-terminally processed, CHLORIDE ION, ... | | 著者 | Marcelot, A, Legrand, P, Zinn-Justin, S. | | 登録日 | 2022-02-25 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.629 Å) | | 主引用文献 | The BAF A12T mutation disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures in Nestor-Guillermo progeria syndrome cells.
Nucleic Acids Res., 50, 2022
|
|
7Z3V
 
 | | Escherichia coli periplasmic phytase AppA D304E mutant, complex with myo-inositol hexakissulfate | | 分子名称: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, POTASSIUM ION | | 著者 | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | 登録日 | 2022-03-02 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
8OSB
 
 | | TWIST1-TCF4-ALX4 complex on specific DNA | | 分子名称: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | 著者 | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | 登録日 | 2023-04-18 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
6WC0
 
 | |
6MTT
 
 | | Crystal structure of VRC46.01 Fab in complex with gp41 peptide | | 分子名称: | Antibody VRC46.01 Fab heavy chain, Antibody VRC46.01 Fb light chain, RV217 founder virus gp41 peptide | | 著者 | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | 登録日 | 2018-10-21 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
5L96
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 3-amino-2-methylpyridine derivative 1 | | 分子名称: | 1,2-ETHANEDIOL, 2-methyl-~{N}-[(2~{R})-1-methylsulfonylpropan-2-yl]pyridin-3-amine, Bromodomain adjacent to zinc finger domain protein 2B | | 著者 | Lolli, G, Marchand, J.-R, Caflisch, A. | | 登録日 | 2016-06-09 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Derivatives of 3-Amino-2-methylpyridine as BAZ2B Bromodomain Ligands: In Silico Discovery and in Crystallo Validation.
J. Med. Chem., 59, 2016
|
|
1A85
 
 | | MMP8 WITH MALONIC AND ASPARAGINE BASED INHIBITOR | | 分子名称: | CALCIUM ION, MMP-8, N~1~-(3-aminobenzyl)-N~2~-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-aspartamide, ... | | 著者 | Brandstetter, H, Roedern, E.G.V, Grams, F, Engh, R.A. | | 登録日 | 1998-04-03 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of malonic acid-based inhibitors bound to human neutrophil collagenase. A new binding mode explains apparently anomalous data.
Protein Sci., 7, 1998
|
|
5I6O
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 20.70 MGy | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | 著者 | Horrell, S, Hough, M.A, Strange, R.W. | | 登録日 | 2016-02-16 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
3L3C
 
 | | Crystal structure of the Bacillus anthracis glmS ribozyme bound to Glc6P | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | 著者 | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | 登録日 | 2009-12-16 | | 公開日 | 2009-12-29 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
5YW1
 
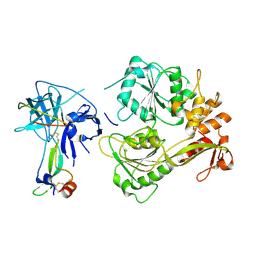 | |
7RU7
 
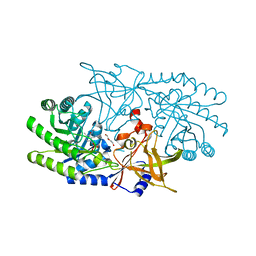 | | Crystal structure of BtrK, a decarboxylase involved in butirosin biosynthesis | | 分子名称: | DI(HYDROXYETHYL)ETHER, L-glutamyl-[BtrI acyl-carrier protein] decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Arenas, L.A.R, Paiva, F.C.R, Huang, F, Leadlay, P, Dias, M.V.B. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of BtrK, a decarboxylase involved in the (S)-4-amino-2-hydroxybutyrate (AHBA) formation during butirosin biosynthesis
J.Mol.Struct., 1267, 2022
|
|
1A6H
 
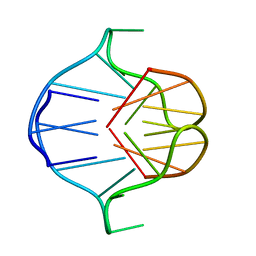 | |
7KZ1
 
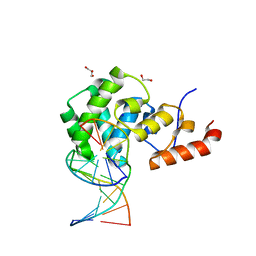 | | Human MBD4 glycosylase domain bound to DNA containing an abasic site | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*CP*GP*(ORP)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), ... | | 著者 | Pidugu, L.S, Bright, H, Pozharski, E, Drohat, A.C. | | 登録日 | 2020-12-09 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
5YWL
 
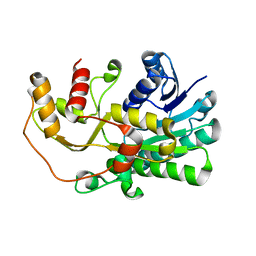 | | SsCR_L211H | | 分子名称: | Protein induced by osmotic stress | | 著者 | Shang, Y.P, Chen, Q, Li, A.T, Yu, H.L, Xu, J.H. | | 登録日 | 2017-11-29 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
7Z71
 
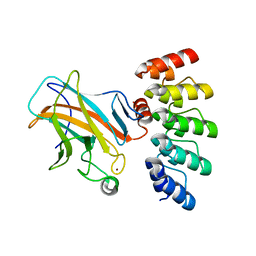 | | Crystal structure of p63 DBD in complex with darpin C14 | | 分子名称: | Darpin C14, Isoform 4 of Tumor protein 63, ZINC ION | | 著者 | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2022-03-14 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
6MVD
 
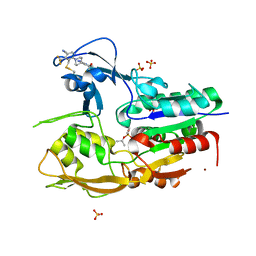 | | Crystal structure of Lecithin:cholesterol acyltransferase (LCAT) in complex with isopropyl dodec-11-enylfluorophosphonate (IDFP) and a small molecule activator | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-{4-[(4R)-4-hydroxy-6-oxo-4-(trifluoromethyl)-4,5,6,7-tetrahydro-2H-pyrazolo[3,4-b]pyridin-3-yl]piperidin-1-yl}-4-(trifluoromethyl)pyridine-3-carbonitrile, NICKEL (II) ION, ... | | 著者 | Manthei, K.A, Chang, L, Tesmer, J.J.G. | | 登録日 | 2018-10-25 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Molecular basis for activation of lecithin:cholesterol acyltransferase by a compound that increases HDL cholesterol.
Elife, 7, 2018
|
|
1AJS
 
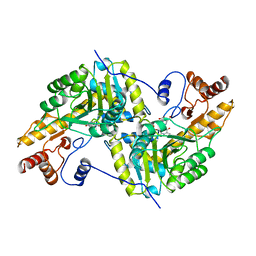 | | REFINEMENT AND COMPARISON OF THE CRYSTAL STRUCTURES OF PIG CYTOSOLIC ASPARTATE AMINOTRANSFERASE AND ITS COMPLEX WITH 2-METHYLASPARTATE | | 分子名称: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, ASPARTATE AMINOTRANSFERASE | | 著者 | Rhee, S, Silva, M.M, Hyde, C.C, Rogers, P.H, Metzler, C.M, Metzler, D.E, Arnone, A. | | 登録日 | 1997-05-08 | | 公開日 | 1997-08-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate.
J.Biol.Chem., 272, 1997
|
|
8P4K
 
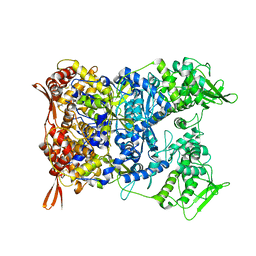 | | Vaccinia Virus palisade layer A10 trimer | | 分子名称: | Core protein OPG136 | | 著者 | Datler, J, Hansen, J.M, Thader, A, Schloegl, A, Hodirnau, V.V, Schur, F.K.M. | | 登録日 | 2023-05-22 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-07-31 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Multi-modal cryo-EM reveals trimers of protein A10 to form the palisade layer in poxvirus cores.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5DOX
 
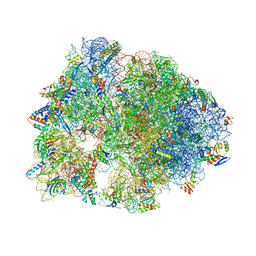 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | 登録日 | 2015-09-11 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
5Z1M
 
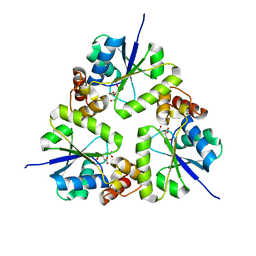 | | Crystal structure of the complex of trimeric Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with citrate ion at 1.87 A resolution | | 分子名称: | CITRIC ACID, Phosphopantetheine adenylyltransferase | | 著者 | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2017-12-26 | | 公開日 | 2018-02-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Crystal structure of the complex of trimeric Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with citrate ion at 1.87 A resolution
To Be Published
|
|
