7A6U
 
 | | Cryo-EM structure of the cytoplasmic domain of human TRPC6 | | 分子名称: | Short transient receptor potential channel 6, UNKNOWN ATOM OR ION | | 著者 | Grieben, M, Pike, A.C.W, Wang, D, Mukhopadhyay, S.M.M, Chalk, R, Marsden, B.D, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2020-08-26 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Cryo-EM structure of the cytoplasmic domain of human TRPC6
TO BE PUBLISHED
|
|
8VLC
 
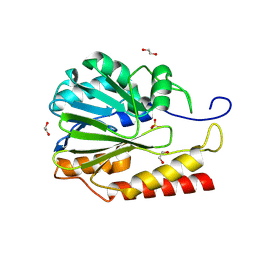 | | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2 | | 分子名称: | 1,2-ETHANEDIOL, HARLDQ motif MBL-fold protein, SULFATE ION | | 著者 | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases, Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2024-01-11 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of Zn-dependent hydrolase from Salmonella typhimurium LT2
To Be Published
|
|
6MEZ
 
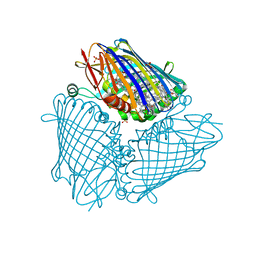 | | X-ray structure of the Fenna-Matthews-Olsen antenna complex from Prosthecochloris aestuarii | | 分子名称: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein, SULFATE ION | | 著者 | Selvaraj, B, Lu, X, Cuneo, M.J, Myles, D.A.A. | | 登録日 | 2018-09-07 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Neutron and X-ray analysis of the Fenna-Matthews-Olson photosynthetic antenna complex from Prosthecochloris aestuarii.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
7TLO
 
 | |
6SOO
 
 | |
7MQV
 
 | | Crystal structure of truncated (ACT domain removed) prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD | | 分子名称: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Shabalin, I.G, Gritsunov, A, Gabryelska, A, Czub, M.P, Grabowski, M, Cooper, D.R, Christendat, D, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-06 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The crystal structure of Bacillus anthracis prephenate dehydrogenase identified an ACT regulatory domain and a novel mode of metabolic regulation for proteins within the prephenate dehydrogenase family of enzyme
to be published
|
|
6ZWV
 
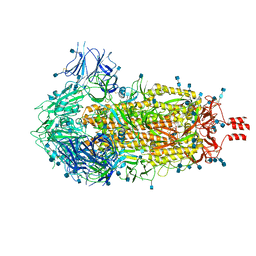 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: 3 Closed RBDs | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Ke, Z, Qu, K, Nakane, T, Xiong, X, Cortese, M, Zila, V, Scheres, S.H.W, Briggs, J.A.G. | | 登録日 | 2020-07-28 | | 公開日 | 2020-08-05 | | 最終更新日 | 2020-12-30 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structures and distributions of SARS-CoV-2 spike proteins on intact virions.
Nature, 588, 2020
|
|
7TIF
 
 | |
6B8G
 
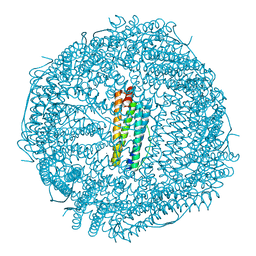 | | Twice-Contracted Human Heavy-Chain Ferritin Crystal-Hydrogel Hybrid | | 分子名称: | CALCIUM ION, FE (III) ION, Ferritin heavy chain | | 著者 | Zhang, L, Bailey, J.B, Subramanian, R, Tezcan, F.A. | | 登録日 | 2017-10-07 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Hyperexpandable, self-healing macromolecular crystals with integrated polymer networks.
Nature, 557, 2018
|
|
8BPR
 
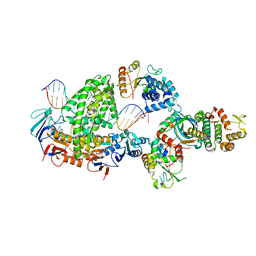 | | Complex of RecF-RecO-RecR-DNA from Thermus thermophilus (low resolution reconstruction). | | 分子名称: | DNA repair protein RecO, DNA replication and repair protein RecF, MAGNESIUM ION, ... | | 著者 | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | 登録日 | 2022-11-17 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7AD9
 
 | | Structure of the Lifeact-F-actin complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Belyy, A, Merino, F, Sitsel, O, Raunser, S. | | 登録日 | 2020-09-14 | | 公開日 | 2020-10-28 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the Lifeact-F-actin complex.
Plos Biol., 18, 2020
|
|
4YH4
 
 | | Crystal structure of human BRD4(1) in complex with 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one (compound 19d) | | 分子名称: | 4-[(5-phenylpyridin-3-yl)carbonyl]-3,4-dihydroquinoxalin-2(1H)-one, Bromodomain-containing protein 4, GLYCEROL, ... | | 著者 | Lakshminarasimhan, D, White, A, Suto, R.K. | | 登録日 | 2015-02-26 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Discovery of a new chemical series of BRD4(1) inhibitors using protein-ligand docking and structure-guided design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5FA9
 
 | | Bifunctional Methionine Sulfoxide Reductase AB (MsrAB) from Treponema denticola | | 分子名称: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptide methionine sulfoxide reductase MsrA | | 著者 | Han, A, Son, J, Kim, H.-Y, Hwang, K.Y. | | 登録日 | 2015-12-11 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Essential Role of the Linker Region in the Higher Catalytic Efficiency of a Bifunctional MsrA-MsrB Fusion Protein
Biochemistry, 55, 2016
|
|
4TQ9
 
 | | Crystal Structure of a GDP-bound G12V Oncogenic Mutant of Human GTPase KRas | | 分子名称: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | 登録日 | 2014-06-10 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.491 Å) | | 主引用文献 | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
5FFL
 
 | |
7PDO
 
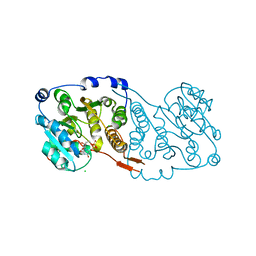 | |
5J1E
 
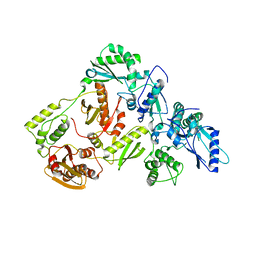 | | Crystal Structure of a Hydroxypyridone Carboxylic Acid Active-Site RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | 分子名称: | 5-hydroxy-4-oxo-1-[(4'-sulfamoyl[1,1'-biphenyl]-4-yl)methyl]-1,4-dihydropyridine-3-carboxylic acid, HIV-1 REVERSE TRANSCRIPTASE P51 DOMAIN, HIV-1 REVERSE TRANSCRIPTASE P66 DOMAIN, ... | | 著者 | Kirby, K.A, Sarafianos, S.G. | | 登録日 | 2016-03-29 | | 公開日 | 2016-06-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H.
J.Med.Chem., 59, 2016
|
|
4DUZ
 
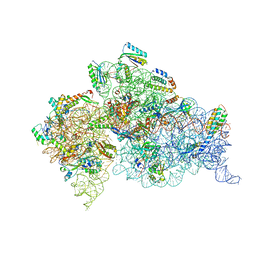 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit with a 16S rRNA mutation, U13C, bound with streptomycin | | 分子名称: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | 著者 | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | 登録日 | 2012-02-22 | | 公開日 | 2013-02-27 | | 実験手法 | X-RAY DIFFRACTION (3.651 Å) | | 主引用文献 | A structural basis for streptomycin resistance
To be Published
|
|
4YPT
 
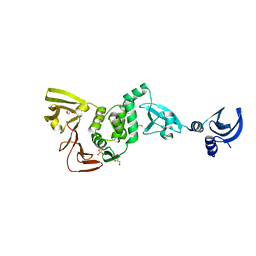 | | X-ray structural of three tandemly linked domains of nsp3 from murine hepatitis virus at 2.60 Angstroms resolution | | 分子名称: | GLYCEROL, Replicase polyprotein 1ab, ZINC ION | | 著者 | Chen, Y, Savinov, S.N, Mielech, A.M, Cao, T, Baker, S.C, Mesecar, A.D. | | 登録日 | 2015-03-13 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.6009 Å) | | 主引用文献 | X-ray Structural and Functional Studies of the Three Tandemly Linked Domains of Non-structural Protein 3 (nsp3) from Murine Hepatitis Virus Reveal Conserved Functions.
J.Biol.Chem., 290, 2015
|
|
6VLH
 
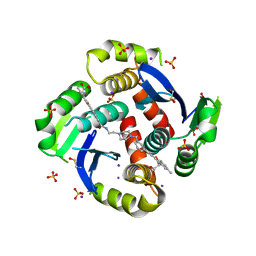 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | 分子名称: | (2-{[3-(4-{2-[(3-{[3-(carboxymethyl)-5-methyl-1-benzofuran-2-yl]ethynyl}benzene-1-carbonyl)amino]ethyl}piperazine-1-carbonyl)phenyl]ethynyl}-5-methyl-1-benzofuran-3-yl)acetic acid, IODIDE ION, Integrase, ... | | 著者 | Gorman, M.A, Parker, M.W. | | 登録日 | 2020-01-24 | | 公開日 | 2021-01-27 | | 実験手法 | X-RAY DIFFRACTION (2.036 Å) | | 主引用文献 | HIV Integrase core domain (IN) in complex with dimeric spanning inhibitor
To Be Published
|
|
4YQ0
 
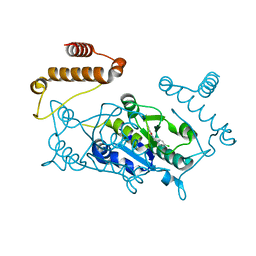 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | 分子名称: | 4-amino-N-(4-chlorobenzyl)-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | 著者 | Elkins, P.A, Bonnette, W.G, Stuckey, J.A. | | 登録日 | 2015-03-13 | | 公開日 | 2016-03-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To be published
|
|
6SYC
 
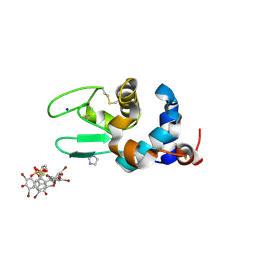 | | Crystal structure of the lysozyme in presence of bromophenol blue at pH 6.5 | | 分子名称: | CHLORIDE ION, IMIDAZOLE, Lysozyme, ... | | 著者 | Camara-Artigas, A, Plaza-Garrido, M, Salinas-Garcia, M.C. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Lysozyme crystals dyed with bromophenol blue: where has the dye gone?
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5FGB
 
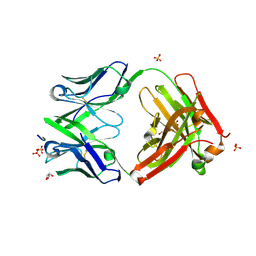 | | Three dimensional structure of broadly neutralizing human anti - Hepatitis C virus (HCV) glycoprotein E2 Fab fragment HC33.4 | | 分子名称: | Anti-HCV E2 Fab HC84-1 heavy chain, Anti-HCV E2 Fab HC84-1 light chain, GLYCEROL, ... | | 著者 | Girard-Blanc, C, Rey, F.A, Krey, T. | | 登録日 | 2015-12-20 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Antibody Response to Hypervariable Region 1 Interferes with Broadly Neutralizing Antibodies to Hepatitis C Virus.
J.Virol., 90, 2016
|
|
6SS5
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0020187 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6FJY
 
 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | 分子名称: | CsuC, Protein CsuE | | 著者 | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | 登録日 | 2018-01-23 | | 公開日 | 2018-05-16 | | 最終更新日 | 2018-09-19 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
