2W3L
 
 | | Crystal Structure of Chimaeric Bcl2-xL and Phenyl Tetrahydroisoquinoline Amide Complex | | Descriptor: | 1-(2-{[(3S)-3-(aminomethyl)-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}phenyl)-4-chloro-5-methyl-N,N-diphenyl-1H-pyrazole-3-carboxamide, APOPTOSIS REGULATOR BCL-2 | | Authors: | Porter, J, Payne, A, de Candole, B, Ford, D, Hutchinson, B, Trevitt, G, Turner, J, Edwards, C, Watkins, C, Whitcombe, I, Davis, J, Stubberfield, C, Fisher, M, Lamers, M. | | Deposit date: | 2008-11-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetrahydroisoquinoline Amide Substituted Phenyl Pyrazoles as Selective Bcl-2 Inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FF7
 
 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | ACETIC ACID, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
6AH9
 
 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and Triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and Triclosan
To Be Published
|
|
1D46
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-100 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
8GC1
 
 | |
5ISQ
 
 | | Staphylococcus aureus H30N, F98Y Dihydrofolate Reductase mutant complexed with beta-NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
4Y66
 
 | |
4LL2
 
 | | Crystal structure of plant lectin with two metal binding sites from cicer arietinum at 2.6 angstrom resolution | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Lectin, ... | | Authors: | Kumar, S, Dube, D, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure plant lectinwith two metal binding sites from cicer arietinum at 2.6 angstrom resolution
TO BE PUBLISHED
|
|
7ACP
 
 | | Serial synchrotron structure of dehaloperoxidase B | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno Chicano, T, Ebrahim, A.E, Axford, D.A, Sherrell, D.A, Worrall, J.W, Strange, R.W, Owen, R.L, Hough, M.A. | | Deposit date: | 2020-09-11 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SFX structure of dehaloperoxidase B in the ferric form
to be published
|
|
8DP5
 
 | | Structure of the PEAK3/14-3-3 complex | | Descriptor: | 14-3-3 protein beta/alpha, 14-3-3 protein epsilon, Protein PEAK3, ... | | Authors: | Torosyan, H, Paul, M, Jura, N, Verba, K.A. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.
Nat Commun, 14, 2023
|
|
3FGX
 
 | | Structure of uncharacterised protein rbstp2171 from bacillus stearothermophilus | | Descriptor: | RBSTP2171 | | Authors: | Minasov, G, Filippova, E.V, Shuvalova, L, Moy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Uncharacterised Protein Rbstp2171 from Bacillus Stearothermophilus
To be Published
|
|
5ISP
 
 | | Staphylococcus aureus F98Y Dihydrofolate Reductase mutant complexed with beta-NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
8A5V
 
 | | Crystal structure of the human phosposerine aminotransferase (PSAT) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase, SULFATE ION | | Authors: | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | Deposit date: | 2022-06-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
2WA8
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - The Phe peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, N-END RULE PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
6OBI
 
 | | Remarkable rigidity of the single alpha-helical domain of myosin-VI revealed by NMR spectroscopy | | Descriptor: | Myosin-VI | | Authors: | Barnes, A, Shen, Y, Ying, J, Takagi, Y, Torchia, D.A, Sellers, J, Bax, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Remarkable Rigidity of the Single alpha-Helical Domain of Myosin-VI As Revealed by NMR Spectroscopy.
J.Am.Chem.Soc., 141, 2019
|
|
8GBZ
 
 | | Crystal structure of PC39-55C, an anti-HIV broadly neutralizing antibody | | Descriptor: | DI(HYDROXYETHYL)ETHER, PC39-55C Fab heavy chain, PC39-55C Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
7RMW
 
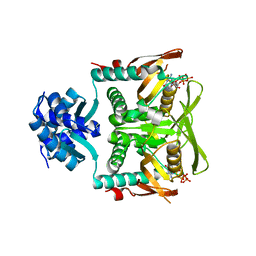 | | Crystal structure of B. subtilis PurR bound to ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Pur operon repressor | | Authors: | Schumacher, M.A. | | Deposit date: | 2021-07-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus.
Nucleic Acids Res., 50, 2022
|
|
5I1R
 
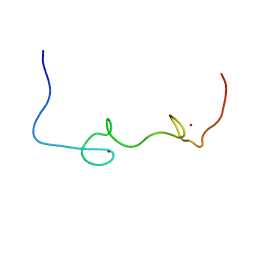 | | Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR and X-ray scattering | | Descriptor: | Nucleocapsid protein p7, ZINC ION | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering.
Chemphyschem, 17, 2016
|
|
3O66
 
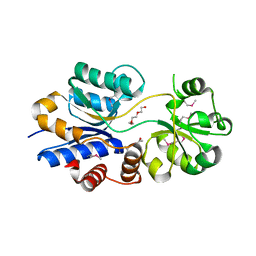 | | Crystal structure of glycine betaine/carnitine/choline ABC transporter | | Descriptor: | ACETATE ION, Glycine betaine/carnitine/choline ABC transporter, TRIETHYLENE GLYCOL | | Authors: | Chang, C, Bigelow, L, Carroll, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of glycine betaine/carnitine/choline ABC transporter
To be Published
|
|
2EKS
 
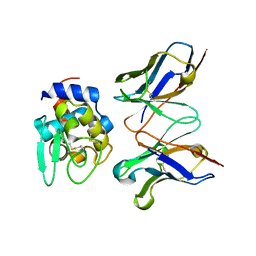 | | Crystal structure of humanized HyHEL-10 FV-HEN lysozyme complex | | Descriptor: | ANTI-LYSOZYME ANTIBODY FV REGION, Lysozyme C | | Authors: | Nakanishi, T, Tsumoto, K, Yokota, A, Kondo, H, Kumagai, I. | | Deposit date: | 2007-03-24 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Critical contribution of VH-VL interaction to reshaping of an antibody: the case of humanization of anti-lysozyme antibody, HyHEL-10
Protein Sci., 17, 2008
|
|
5MLY
 
 | | Closed loop conformation of PhaZ7 Y105E mutant | | Descriptor: | PHB depolymerase PhaZ7 | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
6I80
 
 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT53 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-1-[4-(dimethylamino)piperidin-1-yl]ethanone, Bromodomain-containing protein 2 | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal Structure of the second bromodomain of BRD2 in complex with RT53
To Be Published
|
|
2N49
 
 | | EC-NMR Structure of Erwinia carotovora ECA1580 N-terminal Domain Determined by Combining Evolutionary Couplings (EC) and Sparse NMR Data. Northeast Structural Genomics Consortium target EwR156A | | Descriptor: | Putative cold-shock protein | | Authors: | Tang, Y, Huang, Y.J, Hopf, T.A, Sander, C, Marks, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-06-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination by combining sparse NMR data with evolutionary couplings.
Nat.Methods, 12, 2015
|
|
3OGT
 
 | | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3. | | Descriptor: | (1S,3R,5Z,7E,14beta,17alpha,20S)-20-[5-(1-hydroxy-1-methylethyl)furan-2-yl]-9,10-secopregna-5,7,10-triene-1,3-diol, SULFATE ION, Vitamin D3 receptor | | Authors: | Huet, T, Fraga, R, Mourino, A, Moras, D, Rochel, N. | | Deposit date: | 2010-08-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Chemical synthesis, Functional characterization and Crystal structure of the sidechain analogue of 1,25-dihydroxyvitamin D3.
To be Published
|
|
2ELR
 
 | | Solution structure of the 15th C2H2 zinc finger of human Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 15th C2H2 zinc finger of human Zinc finger protein 406
To be Published
|
|
