6P16
 
 | |
7AXT
 
 | |
7T8V
 
 | | Co-crystal structure of Chaetomium glucosidase I with EB-0159 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T66
 
 | | Co-crystal structure of Chaetomium glucosidase with compound UV-4 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
5MK4
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 7 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[3-(2-methyl-5-phenyl-phenyl)-4-oxidanyl-phenyl]prop-2-enoic acid, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Andrei, S.A, Scheepstra, M, Brunsveld, L, Ottmann, C. | | Deposit date: | 2016-12-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
8CJO
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-06-004 | | Descriptor: | 1-[(3~{S})-3-(4-chloranyl-2-fluoranyl-phenyl)-1,4,8-triazatricyclo[7.4.0.0^{2,7}]trideca-2(7),8-dien-4-yl]-2-(2-ethyl-6-methyl-pyridin-3-yl)oxy-ethanone, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86633706 Å) | | Cite: | Structure-Based Design of Xanthine-Imidazopyridines and -Imidazothiazoles as Highly Potent and In Vivo Efficacious Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 66, 2023
|
|
5MU4
 
 | | Tail Tubular Protein A of Klebsiella pneumoniae bacteriophage KP32 | | Descriptor: | Tail tubular protein A | | Authors: | Pyra, A, Brzozowska, E, Pawlik, K, Dauter, M, Dauter, Z, Gamian, A. | | Deposit date: | 2017-01-12 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tail tubular protein A: a dual-function tail protein of Klebsiella pneumoniae bacteriophage KP32.
Sci Rep, 7, 2017
|
|
8EZS
 
 | | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein | | Descriptor: | CHLORIDE ION, HipS(Lp), HipT(Lp) | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Di Leo, R, Lin, J, Ensminger, A, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of the HipS(Lp)-HipT(Lp) complex from Legionella pneumophila, Sel-met protein
To Be Published
|
|
7T7G
 
 | | Imipenem-OXA-23 2 minute complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T68
 
 | | Co-crystal structure of Chaetomium glucosidase with compound UV-5 | | Descriptor: | (2R,3R,4R,5S)-1-[6-(4-azido-2-nitroanilino)hexyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
7T6W
 
 | | Crystal structure of Chaetomium Glucosidase I (apo) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, GLYCEROL, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Endoplasmic Reticulum alpha-Glucosidase I from a Thermophilic Fungus as a Platform for Structure-Guided Antiviral Drug Design.
Biochemistry, 61, 2022
|
|
6P6L
 
 | | HCV NS3/4A protease domain of genotype 1a in complex with glecaprevir | | Descriptor: | (3aR,7S,10S,12R,21E,24aR)-7-tert-butyl-N-[(1R,2R)-2-(difluoromethyl)-1-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}cyclop ropyl]-20,20-difluoro-5,8-dioxo-2,3,3a,5,6,7,8,11,12,20,23,24a-dodecahydro-1H,10H-9,12-methanocyclopenta[18,19][1,10,17, 3,6]trioxadiazacyclononadecino[11,12-b]quinoxaline-10-carboxamide, 1,2-ETHANEDIOL, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.728 Å) | | Cite: | HCV NS3/4A protease domain of genotype 1a in complex with glecaprevir
To Be Published
|
|
8BZZ
 
 | |
5MWC
 
 | | Crystal structure of the genetically-encoded green calcium indicator NTnC in its calcium bound state | | Descriptor: | CALCIUM ION, genetically-encoded green calcium indicator NTnC | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Rakitina, T.V, Popov, V.O, Subach, O.M, Barykina, N.V, Subach, F.V. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enchanced variant of genetically-encoded green calcium indicator NTnC
To Be Published
|
|
5MWP
 
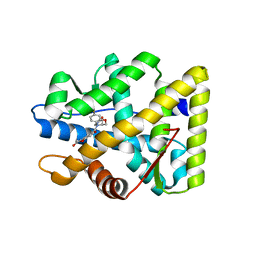 | | The structure of MR in complex with AZD9977. | | Descriptor: | 2-[(3~{S})-7-fluoranyl-4-[(3-oxidanylidene-4~{H}-1,4-benzoxazin-6-yl)carbonyl]-2,3-dihydro-1,4-benzoxazin-3-yl]-~{N}-methyl-ethanamide, Mineralocorticoid receptor, NCOA1 peptide | | Authors: | Edman, K, Aagaard, A, Backstrom, S. | | Deposit date: | 2017-01-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Preclinical pharmacology of AZD9977: A novel mineralocorticoid receptor modulator separating organ protection from effects on electrolyte excretion.
PLoS ONE, 13, 2018
|
|
6USR
 
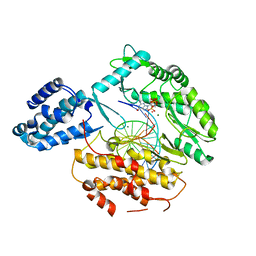 | |
5MWY
 
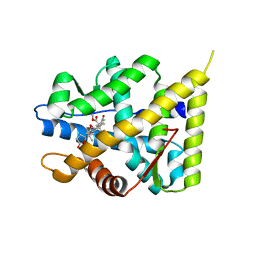 | | The structure of MR in complex with eplerenone. | | Descriptor: | Mineralocorticoid receptor, NCOA1, eplerenone | | Authors: | Edman, K, Aagaard, A, Backstrom, S. | | Deposit date: | 2017-01-20 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Preclinical pharmacology of AZD9977: A novel mineralocorticoid receptor modulator separating organ protection from effects on electrolyte excretion.
PLoS ONE, 13, 2018
|
|
8U4V
 
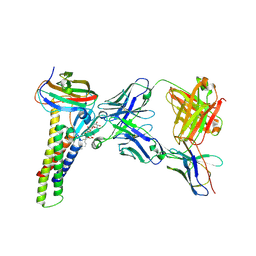 | |
6N23
 
 | |
8C3B
 
 | | X-ray structure of RNase A upon reaction with a Ruthenium(II)-arene Complexed with Glycosylated Carbene Ligands (5) | | Descriptor: | (1,3-dimethyl-2~{H}-imidazol-2-yl)-oxidanyl-oxidanylidene-ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tetrakis(oxidanyl)ruthenium, (1,3-dimethylimidazol-1-ium-2-yl)-tris(oxidanyl)ruthenium, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Ruthenium(II)–Arene Complexes with Glycosylated NHC-Carbene Co-Ligands: Synthesis, Hydrolytic Behavior, and Binding to Biological Molecules
Organometallics, 42, 2023
|
|
8ERN
 
 | | Cyclin-free CDK2 in complex with Cpd21 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, N,3-dimethyl-4-({7-[(2S)-2-methylpyrrolidine-1-carbonyl]quinazolin-2-yl}amino)benzene-1-sulfonamide | | Authors: | Murray, J.M, Oh, A. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cyclin-free CDK2 in complex with Cpd21
To Be Published
|
|
6Y18
 
 | | Ternary complex of 14-3-3 sigma (C38N), Estrogen Related Receptor gamma (DBD) phosphopeptide, and disulfide PPI stabilizer 3 | | Descriptor: | 14-3-3 protein sigma, 4-fluoranyl-~{N}-methyl-~{N}-(2-sulfanylethyl)benzamide, CHLORIDE ION, ... | | Authors: | Somsen, B.A, Ottmann, C. | | Deposit date: | 2020-02-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Acs Chem.Biol., 15, 2020
|
|
7T7D
 
 | | MA-1-206-OXA-23 30s complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
6P6A
 
 | | Structure of Mouse Importin alpha - NIT2 NLS peptide complex | | Descriptor: | Importin subunit alpha-1, Nitrogen catabolic enzyme regulatory protein | | Authors: | Bernardes, N.E, Fukuda, C.A, Silva, T.D, Oliveira, H.C, Fontes, M.R.M. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Comparative study of the interactions between fungal transcription factor nuclear localization sequences with mammalian and fungal importin-alpha.
Sci Rep, 10, 2020
|
|
7T7E
 
 | | MA-1-206-OXA-23 3 minute complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
