5N83
 
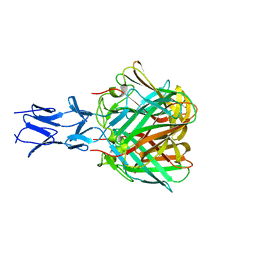 | |
6WGQ
 
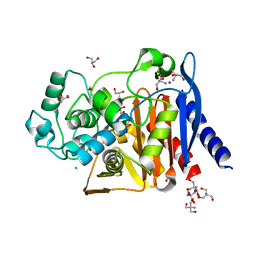 | | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T
To Be Published
|
|
5NRB
 
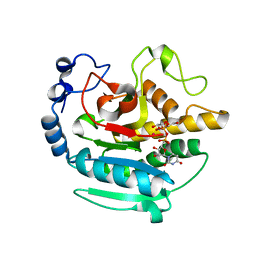 | | A Native Ternary Complex of Alpha-1,3-Galactosyltransferase (a-3GalT) Supports a Conserved Reaction Mechanism for Retaining Glycosyltransferases - alpha-3GalT in complex with Co2+, UDP-Gal and lactose - a3GalT-Co2+-UDP-Gal-LAT-1 | | Descriptor: | COBALT (II) ION, GALACTOSE-URIDINE-5'-DIPHOSPHATE, N-acetyllactosaminide alpha-1,3-galactosyltransferase, ... | | Authors: | Albesa-Jove, D, Marina, A, Sainz-Polo, M.A, Guerin, M.E. | | Deposit date: | 2017-04-22 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Snapshots of alpha-1,3-Galactosyltransferase with Native Substrates: Insight into the Catalytic Mechanism of Retaining Glycosyltransferases.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7OMD
 
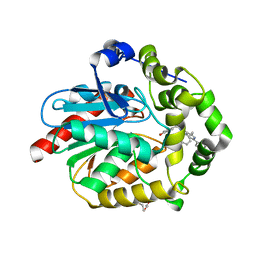 | | Crystal structure of azacoelenterazine-bound Renilla reniformis luciferase variant RLuc8-D162A | | Descriptor: | 6-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)methyl]-8-(phenylmethyl)-[1,2,4]triazolo[4,3-a]pyrazin-3-one, CHLORIDE ION, Coelenterazine h 2-monooxygenase, ... | | Authors: | Schenkmayerova, A, Janin, Y.L, Marek, M. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
6YWM
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
8TZN
 
 | |
7TM3
 
 | | Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
6WNW
 
 | | Active 70S ribosome without free 5S rRNA and bound with A- and P- tRNA | | Descriptor: | 16S ribosomal RNA, 23s-5s joint ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A, Mankin, A.S, Huang, S, Aleksashin, N.A, Klepacki, D, Reier, K, Kefi, A, Szal, A, Remme, J, Jaeger, L, Vazquez-Laslop, N. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly.
Nat Commun, 11, 2020
|
|
8TXP
 
 | | Crystal structure of 05.GC.w13.01 Fab in complex with H1 HA from A/California/04/2009(H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GC_w13_A, Fab heavy chain, ... | | Authors: | Lin, T.H, Moore, N, Wilson, I.A. | | Deposit date: | 2023-08-24 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Maturation of germinal center B cells after influenza virus vaccination in humans.
J.Exp.Med., 221, 2024
|
|
6TPJ
 
 | | Crystal structure of the Orexin-2 receptor in complex with suvorexant at 2.76 A resolution | | Descriptor: | AMMONIUM ION, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Hypocretin receptor-2, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6WT5
 
 | | Structure of a bacterial STING receptor from Capnocytophaga granulosa | | Descriptor: | Bacterial STING | | Authors: | Morehouse, B.R, Govande, A.A, Millman, A, Keszei, A.F.A, Lowey, B, Ofir, G, Shao, S, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2020-05-01 | | Release date: | 2020-09-09 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STING cyclic dinucleotide sensing originated in bacteria.
Nature, 586, 2020
|
|
8TZW
 
 | |
5NCD
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with (2S)-2-amino-5-(diaminomethylideneamino)-N-hydroxypentanamide | | Descriptor: | ACETATE ION, CITRIC ACID, N-HYDROXY-L-ARGININAMIDE, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-03-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
7TUT
 
 | | Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kim, M.K, Lewis, A.J.O, Keenan, R.J, Hegde, R.S. | | Deposit date: | 2022-02-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mechanism of an intramembrane chaperone for multipass membrane proteins.
Nature, 611, 2022
|
|
6Z47
 
 | | Smooth muscle myosin shutdown state heads region | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin heavy chain 11, ... | | Authors: | Scarff, C.A, Carrington, G, Casas Mao, D, Chalovich, J.M, Knight, P.J, Ranson, N.A, Peckham, M. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of the shutdown state of myosin-2.
Nature, 588, 2020
|
|
6QDE
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000877 | | Descriptor: | 3-[[6-~{tert}-butyl-2-[3-(dimethylamino)propyl-methyl-amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6UND
 
 | |
8Q3M
 
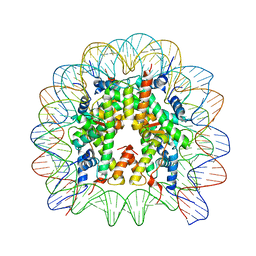 | | Structure of Nucleosome Core with a Bound Kaposi Sarcoma Associated Herpesvirus LANA Peptide Having a Methionine to Ornithine Substitution | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8PWZ
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadBD from Mycobacterium tuberculosis | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadB, UPF0336 protein Rv0504c | | Authors: | Rima, J, Grimoire, Y, Bories, P, Bardou, F, Quemard, A, Bon, C, Mourey, L, Tranier, S. | | Deposit date: | 2023-07-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.00196719 Å) | | Cite: | HadBD dehydratase from Mycobacterium tuberculosis fatty acid synthase type II: A singular structure for a unique function.
Protein Sci., 33, 2024
|
|
6Z5A
 
 | | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941 | | Descriptor: | DIMETHYL SULFOXIDE, N-cyclopentyl-2-[(11,15-dimethyl-10-oxo-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-6-yl)oxy]acetamide, PHOSPHATE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of haspin (GSG2) in complex with macrocycle ODS2002941
To Be Published
|
|
7OSN
 
 | | IRED361 from Micromonospora sp. in complex with NADP+ | | Descriptor: | 6-phosphogluconate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gilio, A.K, Harawa, V, Turner, N, Grogan, G.J. | | Deposit date: | 2021-06-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis of Stereoenriched Piperidines via Chemo-Enzymatic Dearomatization of Activated Pyridines.
J.Am.Chem.Soc., 144, 2022
|
|
7OVO
 
 | | Heterodimeric murine tRNA-guanine transglycosylase in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, Queuine tRNA-ribosyltransferase accessory subunit 2, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
8U08
 
 | |
8GHZ
 
 | | Cryo-EM structure of fish immunogloblin M-Fc | | Descriptor: | Teleost immunoglobulin M protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lyu, M, Stadtmueller, B.M, Malyutin, A.G. | | Deposit date: | 2023-03-13 | | Release date: | 2023-11-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The structure of the teleost Immunoglobulin M core provides insights on polymeric antibody evolution, assembly, and function.
Nat Commun, 14, 2023
|
|
7OR1
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60, conformation 1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grieben, M, Pike, A.C.W, Saward, B.G, Wang, D, Mukhopadhyay, S.M.M, Moreira, T, Chalk, R, MacLean, E.M, Marsden, B.D, Burgess-Brown, N.A, Bountra, C, Schofield, C.J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structure of the human TRPA1 ion channel in complex with the antagonist 3-60
TO BE PUBLISHED
|
|
