8BTU
 
 | |
6YHR
 
 | | Crystal structure of Werner syndrome helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Werner syndrome ATP-dependent helicase, ZINC ION | | Authors: | Newman, J.A, Gavard, A.E, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the helicase core of Werner helicase, a key target in microsatellite instability cancers.
Life Sci Alliance, 4, 2021
|
|
6PYR
 
 | | Human PI3Kdelta in complex with Compound 2-10 ((3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one) | | Descriptor: | (3S)-3-benzyl-3-methyl-5-[5-(2-methylpyrimidin-5-yl)pyrazolo[1,5-a]pyrimidin-3-yl]-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Design of selective PI3K delta inhibitors using an iterative scaffold-hopping workflow.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
8BWB
 
 | | Spider toxin Pha1b (PnTx3-6) from Phoneutria nigriventer targeting CaV2.x calcium channels and TRPA1 channel | | Descriptor: | Omega-ctenitoxin-Pn4a | | Authors: | Mironov, P.A, Chernaya, E.M, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-12-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Recombinant Production, NMR Solution Structure, and Membrane Interaction of the Ph alpha 1 beta Toxin, a TRPA1 Modulator from the Brazilian Armed Spider Phoneutria nigriventer .
Toxins, 15, 2023
|
|
8F5K
 
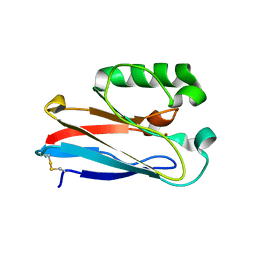 | |
6VJG
 
 | | Csx3-I222 Crystal Form at 1.8 Angstrom Resolution | | Descriptor: | CRISPR-associated protein, Csx3 family | | Authors: | Brown, S, Charbonneau, A, Burman, N, Gauvin, C.C, Lawrence, C.M. | | Deposit date: | 2020-01-15 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA 4 .
J.Biol.Chem., 295, 2020
|
|
8BPO
 
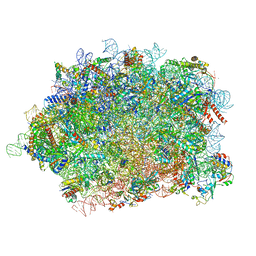 | | Structure of rabbit 80S ribosome translating beta-tubulin in complex with tetratricopeptide protein 5 (TTC5) and S-phase Cyclin A Associated Protein residing in the ER (SCAPER) | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Hopfler, M, Absmeier, E, Passmore, L.A, Hegde, R.S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of ribosome-associated mRNA degradation during tubulin autoregulation.
Mol.Cell, 83, 2023
|
|
8F5L
 
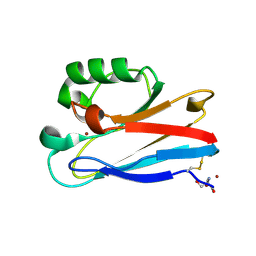 | | Azurin from Pseudomonas aeruginosa, Y72F/Y108F/F110L mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, COPPER (II) ION | | Authors: | Zeug, M, Offenbacher, A.R, Choe, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Electrochemical and Structural Study of the Buried Tryptophan in Azurin: Effects of Hydration and Polarity on the Redox Potential of W48.
J.Phys.Chem.B, 127, 2023
|
|
8EDW
 
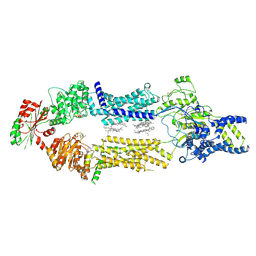 | | Cryo-EM Structure of human ABCA7 in BPL/Ch Nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8BV4
 
 | | Structure of BlaC from Mycobacterium tuberculosis in complex with vaborbactam | | Descriptor: | Beta-lactamase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Chikunova, A, Bruenle, S, Ubbink, M. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Asp179 in the class A beta-lactamase from Mycobacterium tuberculosis is a conserved yet not essential residue due to epistasis.
Febs J., 290, 2023
|
|
6PTE
 
 | | Crystal Structure of ILNAMITKI peptide bound to HLA-A2 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HAUS augmin-like complex subunit 3, ... | | Authors: | Smith, A.R, Arbuiso, A, Keller, G.L.J, Baker, B.M. | | Deposit date: | 2019-07-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure Based Prediction of Neoantigen Immunogenicity.
Front Immunol, 10, 2019
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
8B2S
 
 | | GH24 family muramidase from Trichophaea saccata with an SH3-like cell wall binding domain | | Descriptor: | GH24 family muramidase, POTASSIUM ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5MXX
 
 | | Crystal structure of human SR protein kinase 1 (SRPK1) in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-methyl-~{N}-[2-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]furan-2-carboxamide, ... | | Authors: | Tallant, C, Redondo, C, Batson, J, Toop, H.D, Babaebi-Jadidib, R, Savitsky, P, Elkins, J.M, Newman, J.A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bates, D.O, Morris, J.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Development of Potent, Selective SRPK1 Inhibitors as Potential Topical Therapeutics for Neovascular Eye Disease.
ACS Chem. Biol., 12, 2017
|
|
4YHH
 
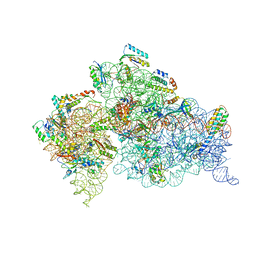 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with tigecycline | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Dhimole, N, Zhou, S, Lopez-Alonso, J.P, Connell, S.R, Fucini, P. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.417 Å) | | Cite: | Structural characterization of an alternative mode of tigecycline binding to the bacterial ribosome.
Antimicrob.Agents Chemother., 59, 2015
|
|
5MOU
 
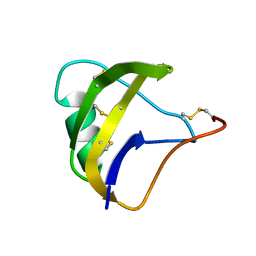 | | NMR spatial structure of scorpion alpha-like toxin BeM9 | | Descriptor: | Alpha-mammal toxin BeM9 | | Authors: | Mineev, K.S, Kuldushev, N.A, Berkut, A.A, Grishin, E.V, Vassilevski, A.A, Arseniev, A.S. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Refined structure of BeM9 reveals arginine hand, an overlooked structural motif in scorpion toxins affecting sodium channels.
Proteins, 86, 2018
|
|
6PVF
 
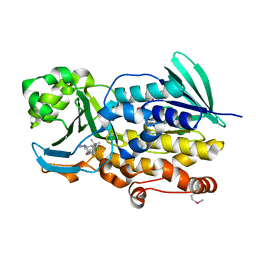 | | Crystal structure of PhqK in complex with malbrancheamide B | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6VLU
 
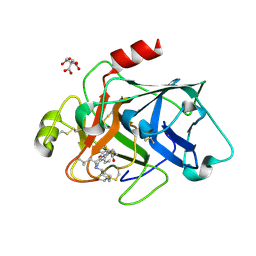 | | Factor XIa in complex with compound 7 | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, N-cyclohexyl-D-leucyl-N-[(1-aminoisoquinolin-6-yl)methyl]-4,4-difluoro-L-prolinamide | | Authors: | Lesburg, C.A, Fradera, X. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-06 | | Last modified: | 2020-10-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Augmenting Hit Identification by Virtual Screening Techniques in Small Molecule Drug Discovery.
J.Chem.Inf.Model., 60, 2020
|
|
6PVH
 
 | | Crystal structure of PhqK in complex with paraherquamide K | | Descriptor: | (7aS,12S,12aR,13aS)-3,3,12,14,14-pentamethyl-3,7,11,12,13,13a,14,15-octahydro-8H,10H-7a,12a-(epiminomethano)indolizino[6,7-h]pyrano[3,2-a]carbazol-16-one, FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
7N6N
 
 | | SARS-CoV-2 Main protease C145S mutant in complex with N and C-terminal residues | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
6PW2
 
 | |
6VOC
 
 | | icosahedral symmetry reconstruction of brome mosaic virus (RNA 3+4) | | Descriptor: | Capsid protein | | Authors: | Beren, C, Cui, Y.X, Chakravarty, A, Yang, X, Rao, A.L.N, Knobler, C.M, Zhou, Z.H, Gelbart, W.M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome organization and interaction with capsid protein in a multipartite RNA virus.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5MBA
 
 | | BINDING MODE OF AZIDE TO FERRIC APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | AZIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1991-01-14 | | Release date: | 1992-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding mode of azide to ferric Aplysia limacina myoglobin. Crystallographic analysis at 1.9 A resolution.
J.Mol.Recog., 4, 1991
|
|
6PVG
 
 | | Crystal structure of ligand free PhqK | | Descriptor: | FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6VP5
 
 | | Ethylene forming enzyme (EFE) D191E variant in complex with Fe(II), L-arginine, and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Davis, K.M, Copeland, R.A, Boal, A.K. | | Deposit date: | 2020-02-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | An Iron(IV)-Oxo Intermediate Initiating l-Arginine Oxidation but Not Ethylene Production by the 2-Oxoglutarate-Dependent Oxygenase, Ethylene-Forming Enzyme.
J.Am.Chem.Soc., 143, 2021
|
|
