3EBC
 
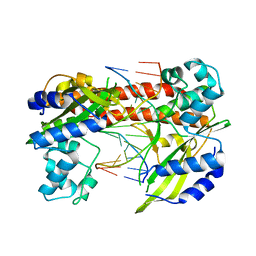 | | Structure of N141A HincII with Cognate DNA | | Descriptor: | 5'-D(*DGP*DCP*DCP*DCP*DGP*DTP*DCP*DGP*DAP*DCP*DCP*DGP*DGP*DC)-3', 5'-D(*DGP*DCP*DCP*DGP*DGP*DTP*DCP*DGP*DAP*DCP*DGP*DGP*DGP*DC)-3', MANGANESE (II) ION, ... | | Authors: | Little, E.J, Babic, A.C, Horton, N.C. | | Deposit date: | 2008-08-27 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Early Interrogation and Recognition of DNA Sequence by Indirect Readout
Structure, 16, 2008
|
|
6IUE
 
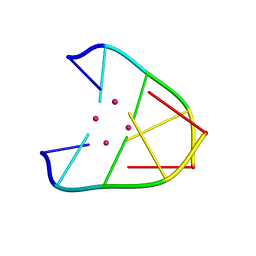 | | DNA helical wire containing Hg(II) | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Ono, A, Kanazawa, H, Ito, H, Goto, M, Nakamura, K, Saneyoshi, H, Kondo, J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A Novel DNA Helical Wire Containing HgII-Mediated T:T and T:G Pairs.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4UWT
 
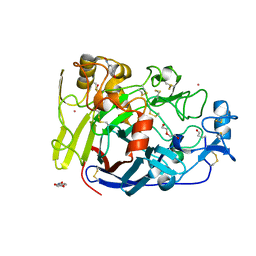 | | Hypocrea jecorina Cel7A E212Q mutant in complex with p-nitrophenyl cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL7A, COBALT (II) ION, ... | | Authors: | Nutt, A, Momeni, M.H, Johansson, G, Stahlberg, J. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 2022
|
|
2HGF
 
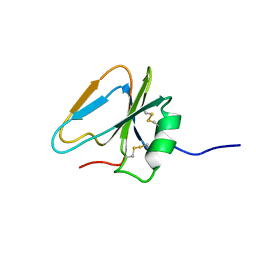 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
1T6H
 
 | | Crystal Structure T4 Lysozyme incorporating an unnatural amino acid p-iodo-L-phenylalanine at position 153 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Spraggon, G, Xie, J, Wang, L, Wu, N, Brock, A, Schultz, P.G. | | Deposit date: | 2004-05-06 | | Release date: | 2004-10-26 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination.
Nat.Biotechnol., 22, 2004
|
|
4QPP
 
 | | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with compound DS-421 (2-{4-[3-CHLORO-2-(2-METHOXYPHENYL)-1H-INDOL-5-YL]PIPERIDIN-1-YL}-N-METHYLETHANAMINE | | Descriptor: | 2-{4-[3-chloro-2-(2-methoxyphenyl)-1H-indol-5-yl]piperidin-1-yl}-N-methylethanamine, POLY-UNK, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, Smil, D, Walker, J.R, He, H, Eram, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Crystal Structure of Human HMT1
hnRNP methyltransferase-like protein 6 in complex with compound DS-421
To be Published
|
|
8A0E
 
 | | CryoEM structure of DHS-eIF5A1 complex | | Descriptor: | Deoxyhypusine synthase, Eukaryotic translation initiation factor 5A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wator, E, Wilk, P, Biela, A.P, Rawski, M, Grudnik, P. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
1T8H
 
 | | 1.8 A CRYSTAL STRUCTURE OF AN UNCHARACTERIZED B. STEAROTHERMOPHILUS PROTEIN | | Descriptor: | BETA-MERCAPTOETHANOL, YlmD protein sequence homologue, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Mondragon, A, Taneja, B, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-12 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A CRYSTAL STRUCTURE OF AN UNCHARACTERIZED B. STEAROTHERMOPHILUS PROTEIN
To be Published
|
|
5CR3
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V104E/L125E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Skerritt, L.A, Bell-Upp, P.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-22 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V104E/L125E at cryogenic temperature
To be Published
|
|
6IEY
 
 | | Crystal structure of Chloramphenicol-Metabolizaing Enzyme EstDL136-Chloramphenicol complex | | Descriptor: | CHLORAMPHENICOL, Esterase | | Authors: | Kim, S.H, Kang, P.A, Han, K.T, Lee, S.W, Rhee, S.K. | | Deposit date: | 2018-09-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome.
PLoS ONE, 14, 2019
|
|
1TFZ
 
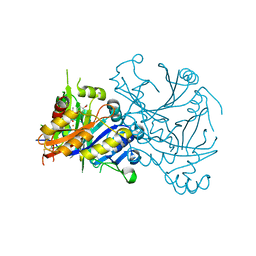 | | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and mammalian 4-hydroxyphenylpyruvate dioxygenases | | Descriptor: | (1-TERT-BUTYL-5-HYDROXY-1H-PYRAZOL-4-YL)[6-(METHYLSULFONYL)-4'-METHOXY-2-METHYL-1,1'-BIPHENYL-3-YL]METHANONE, 4-hydroxyphenylpyruvate dioxygenase, FE (III) ION | | Authors: | Yang, C, Pflugrath, J.W, Camper, D.L, Foster, M.L, Pernich, D.J, Walsh, T.A. | | Deposit date: | 2004-05-27 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for herbicidal inhibitor selectivity revealed by comparison of crystal structures of plant and Mammalian 4-hydroxyphenylpyruvate dioxygenases
Biochemistry, 43, 2004
|
|
3EPV
 
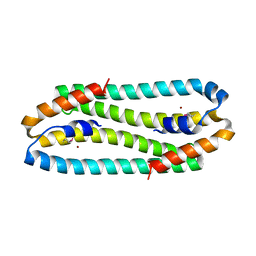 | | X-ray Structure of the Metal-sensor CnrX in both the Apo- and Copper-bound Forms | | Descriptor: | COPPER (II) ION, Nickel and cobalt resistance protein cnrR | | Authors: | Pompidor, G, Maillard, A.P, Girard, E, Gambarelli, S, Kahn, R, Coves, J. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | X-ray structure of the metal-sensor CnrX in both the apo- and copper-bound forms.
Febs Lett., 2008
|
|
6PA4
 
 | |
8I4F
 
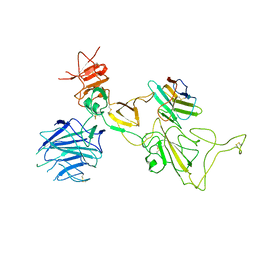 | | Omicron spike variant XBB with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8I4G
 
 | | Omicron spike variant BQ.1.1 with n3130v-Fc | | Descriptor: | Spike glycoprotein, n3130v-Fc | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
6IDR
 
 | | Crystal structure of Vibrio cholerae MATE transporter VcmN in the bent form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MATE family efflux transporter | | Authors: | Kusakizako, T, Claxton, D.P, Tanaka, Y, Maturana, A.D, Kuroda, T, Ishitani, R, Mchaourab, H.S, Nureki, O. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter.
Structure, 27, 2019
|
|
5D55
 
 | | Crystal structure of the E. coli Hda pilus minor tip subunit, HdaB | | Descriptor: | CITRATE ANION, HdaB,HdaA (Adhesin), HUS-associated diffuse adherence, ... | | Authors: | Lee, W.-C, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-08-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and analysis of HdaB: The enteroaggregative Escherichia coli AAF/IV pilus tip protein.
Protein Sci., 25, 2016
|
|
8I4E
 
 | | Omicron spike variant XBB with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
2IH4
 
 | |
3EUW
 
 | |
3EBQ
 
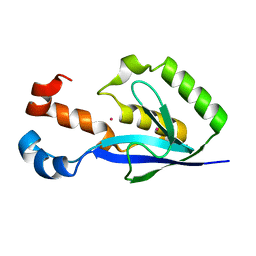 | | Crystal structure of human PPPDE1 | | Descriptor: | MERCURY (II) ION, MOLECULE: PPPDE1 (PERMUTED PAPAIN FOLD PEPTIDASES OF DSRNA VIRUSES AND EUKARYOTES 1), UPF0326 protein FAM152B | | Authors: | Walker, J.R, Akutsu, M, Qiu, L, Li, Y, Slessarev, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Human PPPDE1
To be Published
|
|
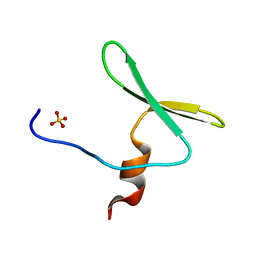
4REX
 
 | |
2IT7
 
 | |
3EB1
 
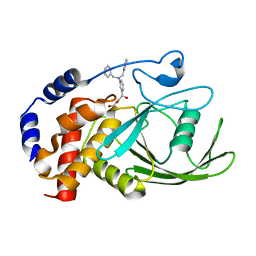 | | Crystal structure PTP1B complex with small molecule inhibitor LZP-25 | | Descriptor: | 4-[3-(dibenzylamino)phenyl]-2,4-dioxobutanoic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Liu, S, Zheng, L.-F, Wu, L, Yu, X, Xue, T, Gunawan, A.M, Long, Y.-Q, Zhang, Z.-Y. | | Deposit date: | 2008-08-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting inactive enzyme conformation: aryl diketoacid derivatives as a new class of PTP1B inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
7ZOM
 
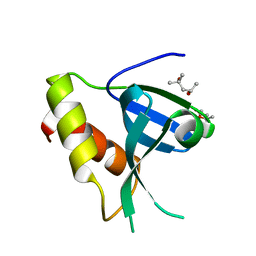 | |
