8OJO
 
 | | Galectin-3 in complex with 2,6-Anhydro-5-S-(beta-D-galactopyranosyl)-5-thio-D-altritol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 1-thio-beta-D-galactopyranose, Galectin-3, ... | | Authors: | Tsagkarakou, A.S, Leonidas, D.D. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strong Binding of C -Glycosylic1,2-Thiodisaccharides to Galectin-3─Enthalpy-Driven Affinity Enhancement by Water-Mediated Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
5DIY
 
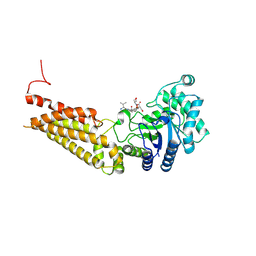 | | Thermobaculum terrenum O-GlcNAc hydrolase mutant - D120N | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronidase, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 | | Authors: | Ostrowski, A, Gundogdu, M, Ferenbach, A.T, Lebedev, A, van Aalten, D.M.F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum.
J.Biol.Chem., 290, 2015
|
|
5K19
 
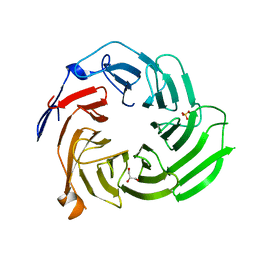 | |
5UNG
 
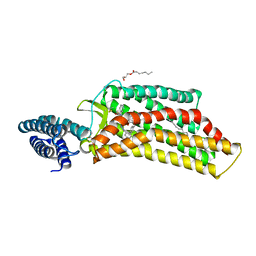 | | XFEL structure of human angiotensin II type 2 receptor (Orthorhombic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl] methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide, ... | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
8C3Q
 
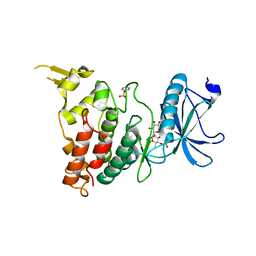 | |
5DII
 
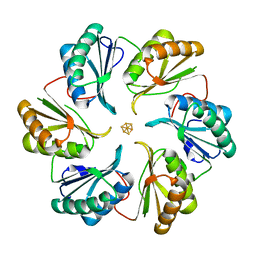 | | Structure of an engineered bacterial microcompartment shell protein binding a [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, Microcompartments protein | | Authors: | Sutter, M, Aussignargues, C, Turmo, A, Kerfeld, C.A. | | Deposit date: | 2015-09-01 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster.
J.Am.Chem.Soc., 138, 2016
|
|
9FJW
 
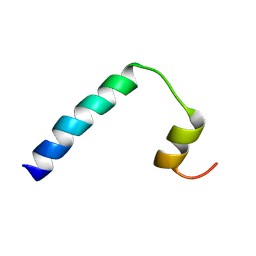 | | Solution NMR structure of a peptide encompassing residues 2-36 of the human formin INF2 | | Descriptor: | Inverted formin-2 | | Authors: | Jimenez, M.A, Comas, L, Labat-de-Hoz, L, Correas, I, Alonso, M.A. | | Deposit date: | 2024-05-31 | | Release date: | 2024-09-11 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the N-terminal extension of the formin INF2.
Cell Mol Life Sci, 79, 2022
|
|
6FZA
 
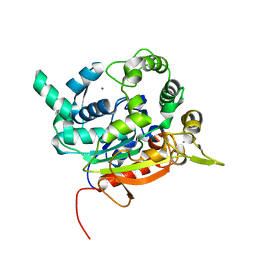 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
4RLG
 
 | |
4TL1
 
 | | GCN4-p1 with mutation to 1-Aminocyclohexanecarboxylic acid at residue 10 | | Descriptor: | General control protein GCN4, ISOPROPYL ALCOHOL | | Authors: | Tavenor, N.A, Silva, K.I, Saxena, S, Horne, W.S. | | Deposit date: | 2014-05-28 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origins of Structural Flexibility in Protein-Based Supramolecular Polymers Revealed by DEER Spectroscopy.
J.Phys.Chem.B, 118, 2014
|
|
4RML
 
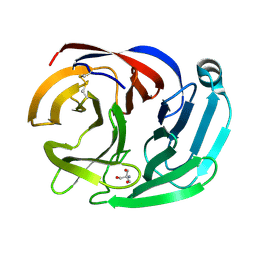 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
4TL6
 
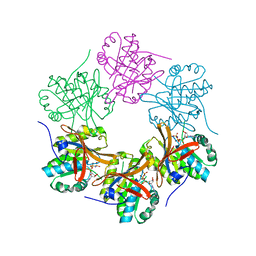 | | Crystal structure of N-terminal domain of KaiC | | Descriptor: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | Authors: | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | Deposit date: | 2014-05-29 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
5DJI
 
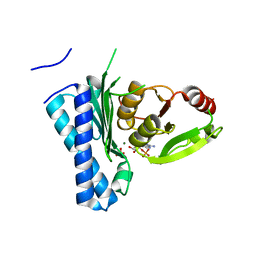 | | Structure of M. tuberculosis CysQ, a PAP phosphatase with AMP, PO4, and 2Mg bound | | Descriptor: | 3'-phosphoadenosine 5'-phosphate phosphatase, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Erickson, A.I. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of Mycobacterium tuberculosis CysQ, with Substrate and Products Bound.
Biochemistry, 54, 2015
|
|
5UQ3
 
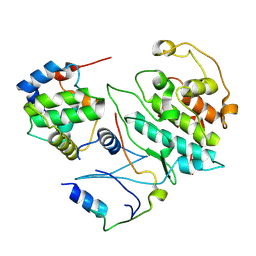 | |
2XJ8
 
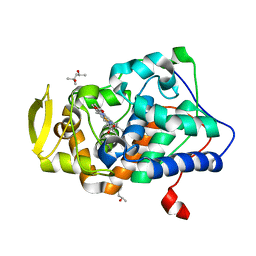 | | The structure of ferrous cytochrome c peroxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2010-07-02 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Nature of the ferryl heme in compounds I and II.
J. Biol. Chem., 286, 2011
|
|
7PP1
 
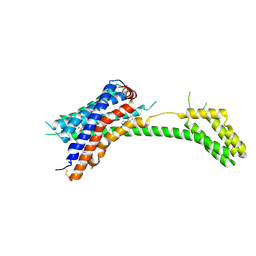 | |
6G0S
 
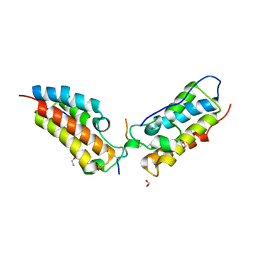 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SIRT7 peptide (K272ac/K275ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, NAD-dependent protein deacetylase sirtuin-7 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
8OYL
 
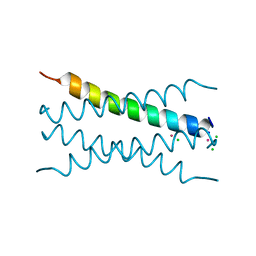 | | Coiled-Coil Domain of Human STIL, Q729L Mutant | | Descriptor: | CADMIUM ION, CHLORIDE ION, SCL-interrupting locus protein, ... | | Authors: | Martin, F.J.O, Shamir, M, Woolfson, D.N, Friedler, A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular Mechanism of STIL Coiled-Coil Domain Oligomerization.
Int J Mol Sci, 24, 2023
|
|
5DQF
 
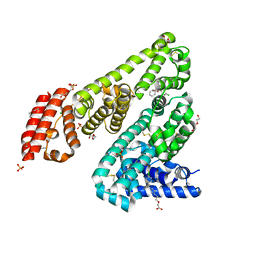 | | Horse Serum Albumin (ESA) in complex with Cetirizine | | Descriptor: | (2-{4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, (2-{4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, CHLORIDE ION, ... | | Authors: | Handing, K.B, Shabalin, I.G, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of equine serum albumin in complex with cetirizine reveals a novel drug binding site.
Mol.Immunol., 71, 2016
|
|
6C5A
 
 | |
8VJ1
 
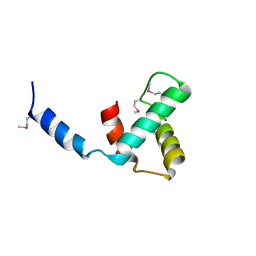 | | Structure of C-terminal domain of telomere resolvase, ResT, from Borrelia garinii | | Descriptor: | Telomere resolvase ResT | | Authors: | Semper, C, Savchenko, A, Watanabe, N, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure analysis of the telomere resolvase from the Lyme disease spirochete Borrelia garinii reveals functional divergence of its C-terminal domain.
Nucleic Acids Res., 52, 2024
|
|
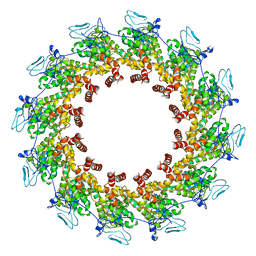
7XPN
 
 | |
3MB2
 
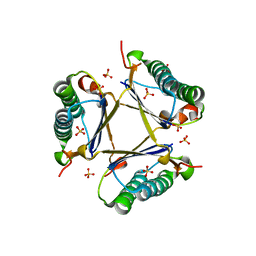 | | Kinetic and Structural Characterization of a Heterohexamer 4-Oxalocrotonate Tautomerase from Chloroflexus aurantiacus J-10-fl: Implications for Functional and Structural Diversity in the Tautomerase Superfamily | | Descriptor: | 4-oxalocrotonate tautomerase family enzyme - alpha subunit, 4-oxalocrotonate tautomerase family enzyme - beta subunit, SULFATE ION | | Authors: | Burks, E.A, Fleming, C.D, Mesecar, A.D, Whitman, C.P, Pegan, S.D. | | Deposit date: | 2010-03-24 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Kinetic and structural characterization of a heterohexamer 4-oxalocrotonate tautomerase from Chloroflexus aurantiacus J-10-fl: implications for functional and structural diversity in the tautomerase superfamily
Biochemistry, 49, 2010
|
|
4RS3
 
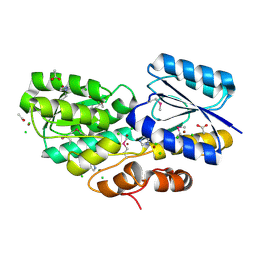 | | Crystal structure of carbohydrate transporter A0QYB3 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with xylitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
6BWC
 
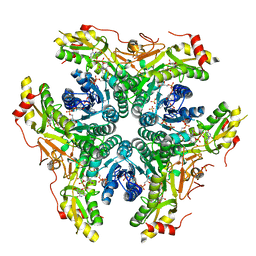 | | X-ray structure of Pen from Bacillus thuringiensis | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Polysaccharide biosynthesis protein CapD, SULFATE ION, ... | | Authors: | Delvaux, N.A, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular architectures of Pen and Pal: Key enzymes required for CMP-pseudaminic acid biosynthesis in Bacillus thuringiensis.
Protein Sci., 27, 2018
|
|
