4S17
 
 | | The crystal structure of glutamine synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, Glutamine synthetase, MAGNESIUM ION | | Authors: | Cuff, M, Tan, K, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-08 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of glutamine synthetase from Bifidobacterium adolescentis ATCC 15703
To be Published
|
|
5L7T
 
 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal inhibitor. | | Descriptor: | (4-fluoranyl-3-oxidanyl-phenyl)-[6-(3-methyl-4-oxidanyl-phenyl)pyridin-2-yl]methanone, 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Braun, F, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | First Structure-Activity Relationship of 17 beta-Hydroxysteroid Dehydrogenase Type 14 Nonsteroidal Inhibitors and Crystal Structures in Complex with the Enzyme.
J. Med. Chem., 59, 2016
|
|
2Q4N
 
 | | Ensemble refinement of the crystal structure of protein from Arabidopsis thaliana At1g79260 | | Descriptor: | Uncharacterized protein At1g79260 | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
6GBR
 
 | | Crystal Structure of the oligomerization domain of VP35 from Reston virus, mercury derivative | | Descriptor: | MERCURIBENZOIC ACID, Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6TFL
 
 | | Lsm protein (SmAP) from Halobacterium salinarum | | Descriptor: | CALCIUM ION, GLYCEROL, RNA-binding protein Lsm, ... | | Authors: | Nikulin, A.D, Fando, M.S, Lekontseva, N.V. | | Deposit date: | 2019-11-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure and RNA-Binding Properties of Lsm Protein from Halobacterium salinarum.
Biochemistry Mosc., 86, 2021
|
|
6TC2
 
 | | Monoclinic human insulin in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Insulin, PHOSPHATE ION, ... | | Authors: | Triandafillidis, D.-P, Parthenios, N, Spiliopoulou, M, Valmas, A, Kosinas, C, Gozzo, F, Reinle-Schmitt, M, Beckers, D, Degen, T, Pop, M, Fitch, A, Wollenhaupt, J, Weiss, M.S, Karavassili, F, Margiolaki, I. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Insulin polymorphism induced by two polyphenols: new crystal forms and advances in macromolecular powder diffraction.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
2B5U
 
 | | Crystal Structure Of Colicin E3 V206C Mutant In Complex With Its Immunity Protein | | Descriptor: | CITRIC ACID, Colicin E3, Colicin E3 immunity protein | | Authors: | Nallini Vijayarangan, A, Nithianantham, S, Nan, W, Jakes, K, Shoham, M. | | Deposit date: | 2005-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Of Colicin E3 In Complex With Its Immunity Protein
TO BE PUBLISHED
|
|
3LEF
 
 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1Z6NA_001 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein 4E10_S0_1Z6NA_001 (T18) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-14 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
5LAR
 
 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
6D13
 
 | | Crystal structure of E.coli RppH-DapF complex | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, IODIDE ION, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2018-04-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF.
Nucleic Acids Res., 46, 2018
|
|
4RG6
 
 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
6G45
 
 | | Crystal structure of mavirus major capsid protein | | Descriptor: | GLYCEROL, Putative major capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6VZY
 
 | |
5VGL
 
 | | Crystal structure of lachrymatory factor synthase from Allium cepa | | Descriptor: | Lachrymatory-factor synthase | | Authors: | Silvaroli, J.A, Pleshinger, M.J, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enzyme That Makes You Cry-Crystal Structure of Lachrymatory Factor Synthase from Allium cepa.
ACS Chem. Biol., 12, 2017
|
|
1TRS
 
 | |
1TT9
 
 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | Descriptor: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | Authors: | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | Deposit date: | 2004-06-22 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
2HW0
 
 | | NMR Solution Structure of the nuclease domain from the Replicator Initiator Protein from porcine circovirus PCV2 | | Descriptor: | Replicase | | Authors: | Vega-Rocha, S, Byeon, I.L, Gronenborn, B, Gronenborn, A.M, Campos-Olivas, R. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2
J.Mol.Biol., 367, 2007
|
|
6W5J
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6CZF
 
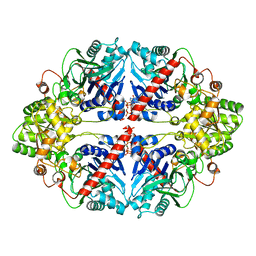 | | The structure of E. coli PurF in complex with ppGpp-Mg | | Descriptor: | Amidophosphoribosyltransferase, GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, B, Grant, R.A, Laub, M.T. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Affinity-based capture and identification of protein effectors of the growth regulator ppGpp.
Nat. Chem. Biol., 15, 2019
|
|
4MYB
 
 | |
1TS3
 
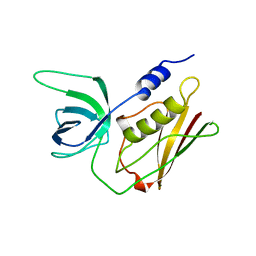 | | H135A MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
6D14
 
 | |
6D2O
 
 | | Beta Carbonic anhydrase in complex with 4-methylimidazole | | Descriptor: | 4-METHYLIMIDAZOLE, Carbonic anhydrase, ZINC ION | | Authors: | Murray, A, Aggarwal, M, Pinard, M, McKenna, R. | | Deposit date: | 2018-04-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mapping of Anion Inhibitors to beta-Carbonic Anhydrase psCA3 from Pseudomonas aeruginosa.
ChemMedChem, 13, 2018
|
|
6IXW
 
 | | pCBH ParM non-polymerisable quadruple mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, pCBH ParM | | Authors: | Koh, F, Popp, D, Narita, A, Robinson, R.C. | | Deposit date: | 2018-12-12 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | The structure of a 15-stranded actin-like filament from Clostridium botulinum.
Nat Commun, 10, 2019
|
|
