8U9J
 
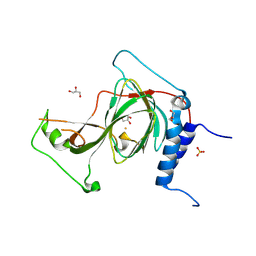 | | The crystal structure of iron-bound human ADO C18S C239S variant at 2.02 Angstrom | | Descriptor: | 2-aminoethanethiol dioxygenase, FE (III) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Duan, R. | | Deposit date: | 2023-09-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Cobalt(II)-Substituted Cysteamine Dioxygenase Oxygenation Proceeds through a Cobalt(III)-Superoxo Complex.
J.Am.Chem.Soc., 146, 2024
|
|
8TG9
 
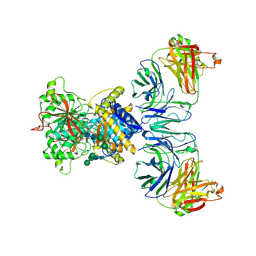 | |
7MVU
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 1-1756; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
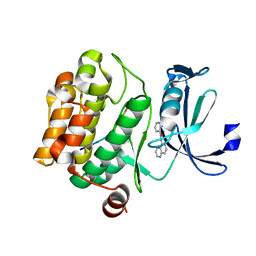
5MZL
 
 | |
4ONG
 
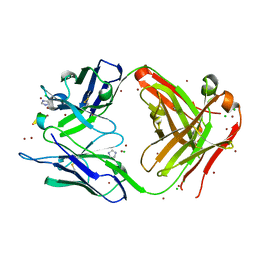 | | Fab fragment of 3D6 in complex with amyloid beta 1-40 | | Descriptor: | 3D6 FAB ANTIBODY HEAVY CHAIN, 3D6 FAB ANTIBODY LIGHT CHAIN, Amyloid beta A4 protein, ... | | Authors: | Feinberg, H, Saldanha, J.W, Diep, L, Goel, A, Widom, A, Veldman, G.M, Weis, W.I, Schenk, D, Basi, G.S. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Alzheimers Res Ther, 6, 2014
|
|
7AD6
 
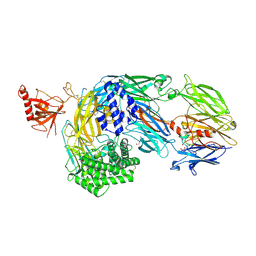 | | Crystal structure of human complement C5 in complex with the K92 bovine knob domain peptide. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CYSTEINE, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
7MVY
 
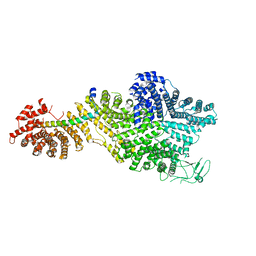 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
6H2F
 
 | | Structure of the pre-pore AhlB of the tripartite alpha-pore forming toxin, AHL, from Aeromonas hydrophila. | | Descriptor: | AhlB, PHOSPHATE ION | | Authors: | Churchill-Angus, A.M, Wilson, J.S, Baker, P.J. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification and structural analysis of the tripartite alpha-pore forming toxin of Aeromonas hydrophila.
Nat Commun, 10, 2019
|
|
7MW0
 
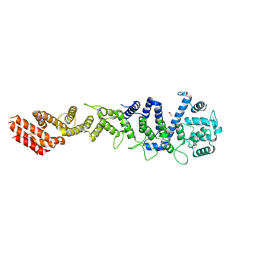 | | Crystal structure of Homo sapiens NUP93 solenoid (residues 174-819) | | Descriptor: | 1,2-ETHANEDIOL, Nuclear pore complex protein Nup93 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MB9
 
 | |
7MB4
 
 | |
7AD7
 
 | | Crystal structure of human complement C5 in complex with the K8 bovine knob domain peptide. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Macpherson, A, van den Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2020-09-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric modulation of Complement C5 by knob domain peptides.
Elife, 10, 2021
|
|
6WJ1
 
 | | Crystal structure of Fab 54-4H03 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-4H03 heavy chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
7MB8
 
 | |
8QUZ
 
 | | Crystal structure of chlorite dismutase at 3000 eV based on analytical absorption corrections | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Duman, R, Wagner, A, Kamps, J, Orville, A. | | Deposit date: | 2023-10-17 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ray-tracing analytical absorption correction for X-ray crystallography based on tomographic reconstructions.
J.Appl.Crystallogr., 57, 2024
|
|
7MB6
 
 | |
7MB7
 
 | |
6OKC
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C12-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N-[2-(dodecanoylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Mindrebo, J.T, Kim, W.E, Bartholow, T.G, Chen, A, Davis, T.D, La Clair, J, Burkart, M.D, Noel, J.P. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Gating mechanism of elongating beta-ketoacyl-ACP synthases.
Nat Commun, 11, 2020
|
|
2IDY
 
 | | NMR Structure of the SARS-CoV non-structural protein nsp3a | | Descriptor: | NSP3 | | Authors: | Serrano, P, Almeida, M.S, Johnson, M.A, Horst, R, Herrmann, T, Joseph, J, Saikatendu, K, Subramanian, V, Stevens, R.C, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus.
J.Virol., 81, 2007
|
|
6C4G
 
 | | Plasmepsin V from Plasmodium vivax bound to a transition state mimetic (WEHI-601) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Czabotar, P.E, Hodder, A.N, Nguyen, W, Sleebs, B.E, Boddey, J.A, Cowman, A.F. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Enhanced antimalarial activity of plasmepsin V inhibitors by modification of the P2position of PEXEL peptidomimetics.
Eur J Med Chem, 154, 2018
|
|
8R2H
 
 | | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Plasmodium falciparum | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gawriljuk, V.O, Godoy, A.S, Oerlemans, R, Groves, M.R. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase DXPS from Plasmodium falciparum reveals a distinct N-terminal domain.
Nat Commun, 15, 2024
|
|
5UE9
 
 | | WT DHODB with orotate bound | | Descriptor: | CHLORIDE ION, Dihydroorotate dehydrogenase, Dihydroorotate dehydrogenase B (NAD(+)), ... | | Authors: | Pompeu, Y.A, Stewart, J.D. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Isoleucine 74 Plays a Key Role in Controlling Electron Transfer Between Lactococcus lactis Dihydroorotate Dehydrogenase 1B Subunits
to be published
|
|
7MB5
 
 | |
7UT3
 
 | | Crystal structure of complex of Fab, G10C with GalNAc-pNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-nitrophenyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside, Fab protein heavy chain, ... | | Authors: | Li, M, Wlodawer, A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Development of a GalNAc-Tyrosine-Specific Monoclonal Antibody and Detection of Tyrosine O -GalNAcylation in Numerous Human Tissues and Cell Lines.
J.Am.Chem.Soc., 144, 2022
|
|
5EUG
 
 | | CRYSTALLOGRAPHIC AND ENZYMATIC STUDIES OF AN ACTIVE SITE VARIANT H187Q OF ESCHERICHIA COLI URACIL DNA GLYCOSYLASE: CRYSTAL STRUCTURES OF MUTANT H187Q AND ITS URACIL COMPLEX | | Descriptor: | PROTEIN (GLYCOSYLASE), URACIL | | Authors: | Xiao, G, Tordova, M, Drohat, A.C, Jagadeesh, J, Stivers, J.T, Gilliland, G.L. | | Deposit date: | 1998-12-27 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Escherichia coli uracil DNA glycosylase and its complexes with uracil and glycerol: structure and glycosylase mechanism revisited.
Proteins, 35, 1999
|
|
