5HGC
 
 | | A Serpin structure | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, SULFATE ION, ... | | Authors: | Das, S, Vashchenko, G.V, Van Petegem, F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | A Serpin structure
J.Biol.Chem., 2016
|
|
3QR5
 
 | |
8F5B
 
 | |
6VSV
 
 | |
6VRR
 
 | |
8SEW
 
 | | Cryo-EM Structure of RyR1 + ADP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SF0
 
 | | Cryo-EM Structure of RyR1 + cAMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEN
 
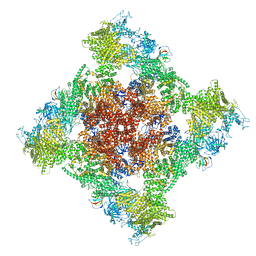 | | Cryo-EM Structure of RyR1 | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEZ
 
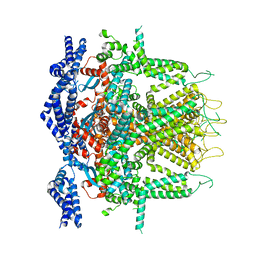 | | Cryo-EM Structure of RyR1 + Adenine (Local Refinement of TMD) | | Descriptor: | ADENINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEV
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S (Local Refinement of TMD) | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SES
 
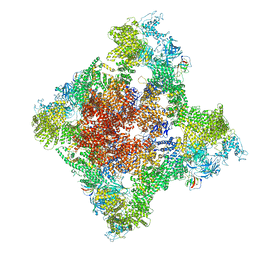 | | Cryo-EM Structure of RyR1 + Adenine | | Descriptor: | ADENINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEP
 
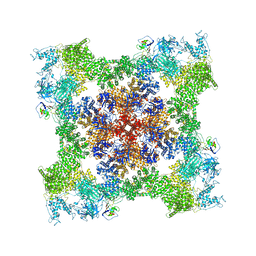 | | Cryo-EM Structure of RyR1 + ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEQ
 
 | | Cryo-EM Structure of RyR1 + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEY
 
 | | Cryo-EM Structure of RyR1 + Adenosine (Local Refinement of TMD) | | Descriptor: | ADENOSINE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SET
 
 | | Cryo-EM Structure of RyR1 + cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEO
 
 | | Cryo-EM Structure of RyR1 + ATP-gamma-S | | Descriptor: | Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEX
 
 | | Cryo-EM Structure of RyR1 + AMP (Local Refinement of TMD) | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SER
 
 | | Cryo-EM Structure of RyR1 + Adenosine | | Descriptor: | ADENOSINE, Glutathione S-transferase class-mu 26 kDa isozyme,Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 1, ... | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
8SEU
 
 | | Cryo-EM Structure of RyR1 (Local Refinement of TMD) | | Descriptor: | Ryanodine receptor 1, ZINC ION | | Authors: | Cholak, S, Saville, J.W, Zhu, X, Berezuk, A.M, Tuttle, K.S, Haji-Ghassemi, O, Van Petegem, F, Subramaniam, S. | | Deposit date: | 2023-04-10 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives.
Structure, 31, 2023
|
|
6PAL
 
 | | Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus | | Descriptor: | ACETATE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tamura, K, Brumer, H, van Petegem, F. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Synergy between Cell Surface Glycosidases and Glycan-Binding Proteins Dictates the Utilization of Specific Beta(1,3)-Glucans by Human GutBacteroides.
Mbio, 11, 2020
|
|
6M2W
 
 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6PYA
 
 | |
6PYF
 
 | | Sex Hormone-binding globulin mutant E176K in complex with Estradiol | | Descriptor: | CALCIUM ION, ESTRADIOL, Sex hormone-binding globulin | | Authors: | Round, P.W, Wu, T.S, Das, S, Van Petegem, F. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Molecular interactions between sex hormone-binding globulin and nonsteroidal ligands that enhance androgen activity.
J.Biol.Chem., 295, 2020
|
|
6PYB
 
 | | Sex Hormone-binding globulin mutant E176K in complex with DVT | | Descriptor: | 4,4'-[(3R,4R)-oxolane-3,4-diylbis(methylene)]bis(2-methoxyphenol), CALCIUM ION, Sex hormone-binding globulin | | Authors: | Round, P.W, Das, S, Van Petegem, F. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2020-02-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular interactions between sex hormone-binding globulin and nonsteroidal ligands that enhance androgen activity.
J.Biol.Chem., 295, 2020
|
|
6MUD
 
 | |
