2PJZ
 
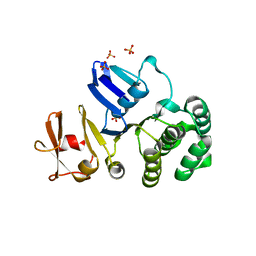 | | The crystal structure of putative Cobalt transport ATP-binding protein (cbiO-2), ST1066 | | Descriptor: | Hypothetical protein ST1066, SULFATE ION | | Authors: | Hirata, K, Hasegawa, K, Ebihara, A, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-17 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of putative Cobalt transport ATP-binding protein (cbiO-2), ST1066
To be Published
|
|
1WUA
 
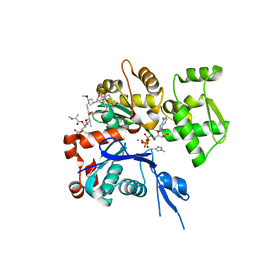 | | The structure of Aplyronine A-actin complex | | Descriptor: | (8R,9R,10R,11R,14S,18S,20S,24S)-24-{(1R,2S,3R,6R,7R,8R,9S,10E)-8-(ACETYLOXY)-6-[(N,N-DIMETHYLALANYL)OXY]-11-[FORMYL(MET HYL)AMINO]-2-HYDROXY-1,3,7,9-TETRAMETHYLUNDEC-10-ENYL}-10-HYDROXY-14,20-DIMETHOXY-9,11,15,18-TETRAMETHYL-2-OXOOXACYCLOTE TRACOSA-3,5,15,21-TETRAEN-8-YL N,N,O-TRIMETHYLSERINATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Hirata, K, Muraoka, S, Suenaga, K, Kuroda, T, Kato, K, Tanaka, H, Yamamoto, M, Takata, M, Yamada, K, Kigoshi, H. | | Deposit date: | 2004-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure basis for antitumor effect of aplyronine a
J.Mol.Biol., 356, 2006
|
|
3WG7
 
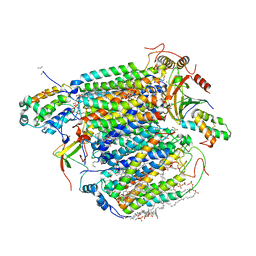 | | A 1.9 angstrom radiation damage free X-ray structure of large (420KDa) protein by femtosecond crystallography | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Hirata, K, Shinzawa-Itoh, K, Yano, N, Takemura, S, Kato, K, Hatanaka, M, Muramoto, K, Kawahara, T, Tsukihara, T, Yamashita, E, Tono, K, Ueno, G, Hikima, T, Murakami, H, Inubushi, Y, Yabashi, M, Ishikawa, T, Yamamoto, M, Ogura, T, Sugimoto, H, Shen, J.R, Yoshikawa, S, Ago, H. | | Deposit date: | 2013-07-29 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
7Y44
 
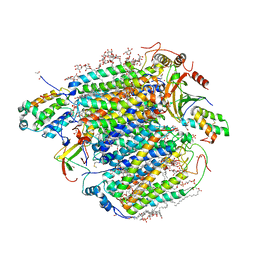 | | Re-refinement of damage free X-ray structure of bovine cytochrome c oxidase at 1.9 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Hirata, K, Ago, H. | | Deposit date: | 2022-06-14 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL.
Nat.Methods, 11, 2014
|
|
5AYG
 
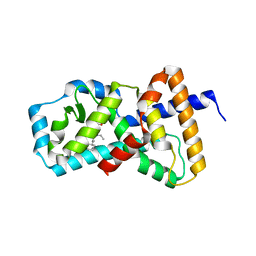 | | Crystal Structure of the Human ROR gamma Ligand Binding Domain With 3g | | Descriptor: | 3-[5-(2-cyclohexylethyl)-4-ethyl-1,2,4-triazol-3-yl]-N-naphthalen-1-yl-propanamide, Nuclear receptor ROR-gamma | | Authors: | Noguchi, M, Doi, S, Nomura, A, Kikuwaka, M, Murase, K, Hirata, K, Kamada, M, Adachi, T. | | Deposit date: | 2015-08-20 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | SAR Exploration Guided by LE and Fsp(3): Discovery of a Selective and Orally Efficacious ROR gamma Inhibitor
Acs Med.Chem.Lett., 7, 2016
|
|
7BU7
 
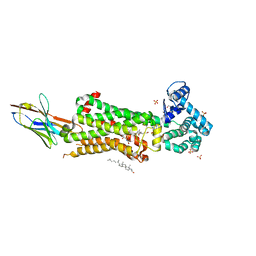 | | Structure of human beta1 adrenergic receptor bound to BI-167107 and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
5GX4
 
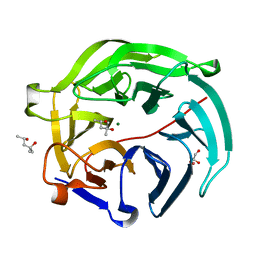 | | Luciferin-regenerating enzyme collected with serial synchrotron rotational crystallography with accumulated dose of 14 MGy (12th measurement) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, Luciferin regenerating enzyme, ... | | Authors: | Hasegawa, K, Yamashita, K, Murai, T, Nuemket, N, Hirata, K, Ueno, G, Ago, H, Nakatsu, T, Kumasaka, T, Yamamoto, M. | | Deposit date: | 2016-09-15 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography
J Synchrotron Radiat, 24, 2017
|
|
8GQ1
 
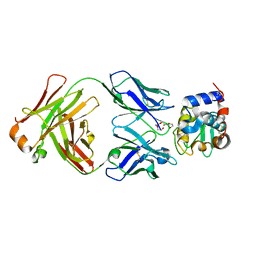 | | HyHEL10 Fab complexed with hen egg lysozyme carrying arginine cluster in framework region of light chain. | | Descriptor: | Heavy Chain of HyHel10 Antibody Fragment (Fab), Light Chain of HyHel10 Antibody Fragment (Fab), Lysozyme C, ... | | Authors: | Matsuura, H, Hirata, K, Sakai, N, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-08-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Arginine cluster introduction on framework region in anti-lysozyme antibody improved association rate constant by changing conformational diversity of CDR loops.
Protein Sci., 32, 2023
|
|
6IGL
 
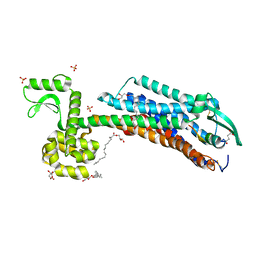 | | Crystal Structure of human ETB receptor in complex with IRL1620 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Endothelin receptor type B,Endolysin,Endothelin receptor type B, ... | | Authors: | Shihoya, W, Izume, T, Inoue, A, Yamashita, K, kadji, F.M.N, Hirata, K, Aoki, J, Nishizawa, T, Nureki, O. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human ETBreceptor provide mechanistic insight into receptor activation and partial activation.
Nat Commun, 9, 2018
|
|
4UB8
 
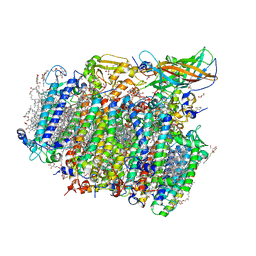 | | Native structure of photosystem II (dataset-2) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
4UB6
 
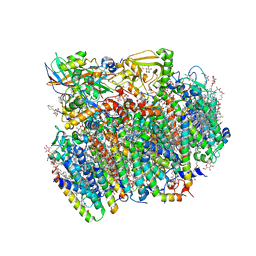 | | Native structure of photosystem II (dataset-1) by a femtosecond X-ray laser | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Akita, F, Hirata, K, Ueno, G, Murakami, H, Nakajima, Y, Shimizu, T, Yamashita, K, Yamamoto, M, Ago, H, Shen, J.R. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Native structure of photosystem II at 1.95 angstrom resolution viewed by femtosecond X-ray pulses.
Nature, 517, 2015
|
|
6E67
 
 | | Structure of beta2 adrenergic receptor fused to a Gs peptide | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor,Endolysin,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Beta-2 adrenergic receptor chimera | | Authors: | Liu, X, Xu, X, Hilger, D, Tiemann, J, Liu, H, Du, Y, Hirata, K, Sun, X, Guixa-Gonzalez, R, Mathiesen, J, Hildebrand, P, Kobilka, B. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
8W7N
 
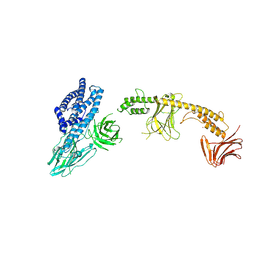 | | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis | | Descriptor: | Pesticidal crystal protein Cry1Aa, UNKNOWN ATOM OR ION | | Authors: | Tanaka, J, Abe, S, Hayakawa, T, Kojima, M, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2023-08-31 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 685, 2023
|
|
6OBA
 
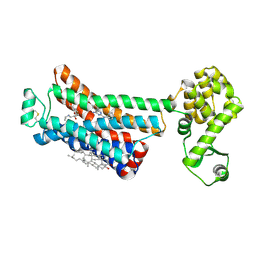 | | The beta2 adrenergic receptor bound to a negative allosteric modulator | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, 6-bromo-N~2~-phenylquinazoline-2,4-diamine, Beta-2 adrenergic receptor,Lysozyme,Beta-2 adrenergic receptor, ... | | Authors: | Liu, X, Stobel, A, Kaindl, J, Dengler, D, ClarK, M, Mahoney, J, Korczynska, M, Matt, R.A, Hubner, H, Xu, X, Stanek, M, Hirata, K, Shoichet, B, Sunahara, R, Gmeiner, R, Kobilka, B.K. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An allosteric modulator binds to a conformational hub in the beta2adrenergic receptor.
Nat.Chem.Biol., 16, 2020
|
|
4YSP
 
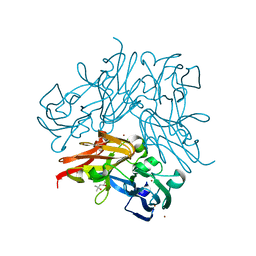 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.32 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diedrichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSD
 
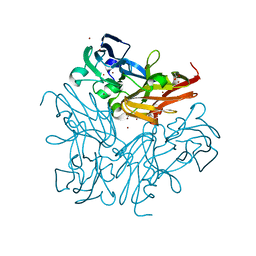 | | Room temperature structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | ACETIC ACID, COPPER (II) ION, Nitrite reductase | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSQ
 
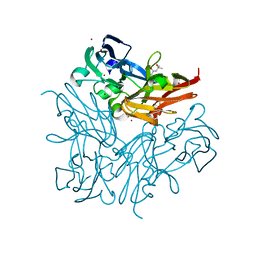 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 8.38 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YST
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSO
 
 | | Copper nitrite reductase from Geobacillus thermodenitrificans - 0.064 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
4YSA
 
 | | Completely oxidized structure of copper nitrite reductase from Geobacillus thermodenitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, SODIUM ION | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
8J2Q
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JAE
 
 | |
8X8S
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WLG
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X8V
 
 | | Crystal structure of Cypovirus Polyhedra mutant fused with c-Myc fragment | | Descriptor: | Polyhedrin,Myc proto-oncogene protein | | Authors: | Kojima, M, Ueno, T, Abe, S, Hirata, K. | | Deposit date: | 2023-11-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-throughput structure determination of an intrinsically disordered protein using cell-free protein crystallization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
