2QR9
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed with an Oxabicyclic Derivative Compound | | 分子名称: | Estrogen receptor, Nuclear receptor coactivator 2, dimethyl (1R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hepta-2,5-diene-2,3-dicarboxylate | | 著者 | Nettles, K.W, Bruning, J.B. | | 登録日 | 2007-07-27 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
4M6T
 
 | | Structure of human Paf1 and Leo1 complex | | 分子名称: | RNA polymerase II-associated factor 1 homolog, Linker, RNA polymerase-associated protein LEO1, ... | | 著者 | Shen, Y, Qin, X. | | 登録日 | 2013-08-11 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | Structural insights into Paf1 complex assembly and histone binding
Nucleic Acids Res., 41, 2013
|
|
2B23
 
 | | Human estrogen receptor alpha ligand-binding domain and a glucocorticoid receptor-interacting protein 1 NR box II peptide | | 分子名称: | Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | 登録日 | 2005-09-16 | | 公開日 | 2006-09-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
8GUO
 
 | |
8GUP
 
 | |
8GUN
 
 | |
5CQR
 
 | | Dimerization of Elp1 is essential for Elongator complex assembly | | 分子名称: | Elongator complex protein 1 | | 著者 | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | 登録日 | 2015-07-22 | | 公開日 | 2015-08-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.015 Å) | | 主引用文献 | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CQS
 
 | | Dimerization of Elp1 is essential for Elongator complex assembly | | 分子名称: | Elongator complex protein 1 | | 著者 | Lin, Z, Xu, H, Li, F, Diao, W, Long, J, Shen, Y. | | 登録日 | 2015-07-22 | | 公開日 | 2015-08-19 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Dimerization of elongator protein 1 is essential for Elongator complex assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QKN
 
 | | Crystal structure of FTO bound to a selective inhibitor | | 分子名称: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | 著者 | Yang, C.-G, Huang, Y, Gan, J. | | 登録日 | 2014-06-07 | | 公開日 | 2014-12-03 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5.
Nucleic Acids Res., 43, 2015
|
|
2ESW
 
 | |
3U3I
 
 | | A RNA binding protein from Crimean-Congo hemorrhagic fever virus | | 分子名称: | Nucleocapsid protein | | 著者 | Guo, Y, Wang, W.M, Ji, W, Deng, M, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | 登録日 | 2011-10-06 | | 公開日 | 2012-03-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.304 Å) | | 主引用文献 | Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7L7Q
 
 | | Ctf3c with Ulp2-KIM | | 分子名称: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | 著者 | Hinshaw, S.M, Harrison, S.C. | | 登録日 | 2020-12-30 | | 公開日 | 2021-02-10 | | 最終更新日 | 2021-08-25 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ctf3/CENP-I provides a docking site for the desumoylase Ulp2 at the kinetochore.
J.Cell Biol., 220, 2021
|
|
6LT8
 
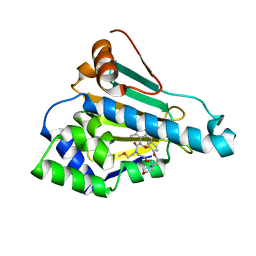 | | HSP90 in complex with KW-2478 | | 分子名称: | 2-[2-ethyl-6-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]carbonyl-3,5-bis(oxidanyl)phenyl]-~{N},~{N}-bis(2-methoxyethyl)ethanamide, Heat shock protein HSP 90-alpha | | 著者 | Cao, H.L. | | 登録日 | 2020-01-21 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.593 Å) | | 主引用文献 | Anti-NSCLC activity in vitro of Hsp90 N inhibitor KW-2478 and complex crystal structure determination of Hsp90 N -KW-2478.
J.Struct.Biol., 213, 2021
|
|
7LQ1
 
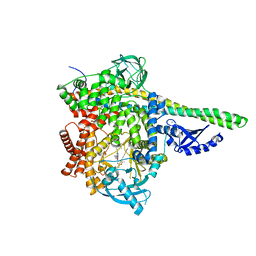 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 28 | | 分子名称: | CHLORIDE ION, N-(5-(6-(2-((2S,6R)-2,6-dimethylmorpholino)pyridin-4-yl)-1-oxoisoindolin-4-yl)-2-methoxypyridin-3-yl)methanesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | 著者 | Lesburg, C.A, Augustin, M. | | 登録日 | 2021-02-12 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.96 Å) | | 主引用文献 | Discovery of a new series of PI3K-delta inhibitors from Virtual Screening.
Bioorg.Med.Chem.Lett., 42, 2021
|
|
6LR9
 
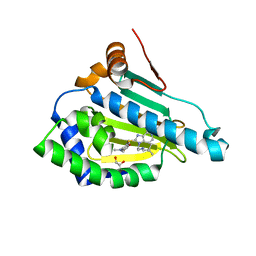 | | HSP90 in complex with Debio0932 | | 分子名称: | 2-[[6-(dimethylamino)-1,3-benzodioxol-5-yl]sulfanyl]-1-[2-(2,2-dimethylpropylamino)ethyl]imidazo[4,5-c]pyridin-4-amine, GLYCEROL, Heat shock protein HSP 90-alpha | | 著者 | Cao, H.L. | | 登録日 | 2020-01-15 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.196 Å) | | 主引用文献 | Complex crystal structure determination and anti-non-small-cell lung cancer activity of the Hsp90 N inhibitor Debio0932.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8T0P
 
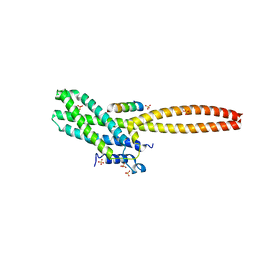 | | Structure of Cse4 bound to Ame1 and Okp1 | | 分子名称: | Histone H3-like centromeric protein CSE4, Inner kinetochore subunit AME1, Inner kinetochore subunit OKP1, ... | | 著者 | Deng, S, Harrison, S.C. | | 登録日 | 2023-06-01 | | 公開日 | 2023-09-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Recognition of centromere-specific histone Cse4 by the inner kinetochore Okp1-Ame1 complex.
Embo Rep., 24, 2023
|
|
5VKU
 
 | | An atomic structure of the human cytomegalovirus (HCMV) capsid with its securing layer of pp150 tegument protein | | 分子名称: | Major capsid protein, Small capsomere-interacting protein, Tegument protein pp150, ... | | 著者 | Yu, X, Jih, J, Jiang, J, Zhou, H. | | 登録日 | 2017-04-24 | | 公開日 | 2017-06-28 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150.
Science, 356, 2017
|
|
4FF5
 
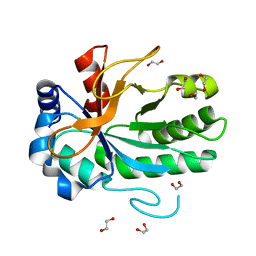 | |
3TUO
 
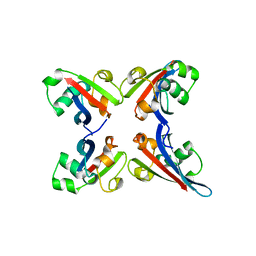 | |
3UIT
 
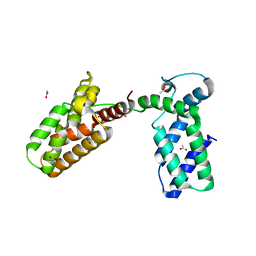 | | Overall structure of Patj/Pals1/Mals complex | | 分子名称: | ACETATE ION, InaD-like protein, MAGUK p55 subfamily member 5, ... | | 著者 | Zhang, J, Yang, X, Long, J, Shen, Y. | | 登録日 | 2011-11-06 | | 公開日 | 2012-02-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of an L27 domain heterotrimer from cell polarity complex Patj/Pals1/Mals2 reveals mutually independent L27 domain assembly mode
J.Biol.Chem., 287, 2012
|
|
8H5F
 
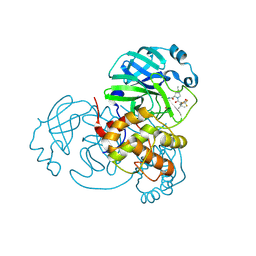 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) L167F Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H4Y
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6N
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (T21I) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-18 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7K
 
 | |
8H5P
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
