8QVW
 
 | | Cryo-EM structure of the peptide binding domain of human SRP68/72 | | Descriptor: | Signal recognition particle subunit SRP68, Signal recognition particle subunit SRP72 | | Authors: | Zhong, Y, Feng, J, Koh, A.F, Kotecha, A, Greber, B.J, Ataide, S.F. | | Deposit date: | 2023-10-18 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of nPBD of human SRP68/72
To Be Published
|
|
2LSP
 
 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
5JOM
 
 | | X-ray structure of CO-bound sperm whale myoglobin using a fixed target crystallography chip | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Oghbaey, S, Sarracini, A, Ginn, H.M, Pare-Labrosse, O, Kuo, A, Marx, A, Epp, S.W, Sherrell, D.A, Eger, B.T, Zhong, Y, Loch, R, Mariani, V, Alonso-Mori, R, Nelson, S, Lemke, H.T, Owen, R.L, Pearson, A.R, Stuart, D.I, Ernst, O.P, Mueller-Werkmeister, H.M, Miller, R.J.D. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8JMT
 
 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | Descriptor: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | Authors: | Tao, Y, Guo, Q, He, B, Zhong, Y. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
7DMC
 
 | | Dipyridamole binds to the N-terminal domain of human Hsp90A | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, CHLORIDE ION, Heat shock protein HSP 90-alpha, ... | | Authors: | Shi, L, Zhou, C, Zhong, Y, Gao, J, Zhou, H, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dipyridamole interacts with the N-terminal domain of HSP90 and antagonizes the function of the chaperone in multiple cancer cell lines.
Biochem Pharmacol, 207, 2022
|
|
8D4Y
 
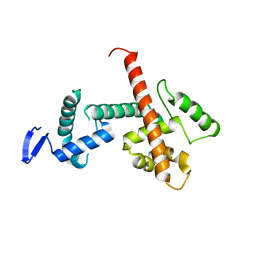 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
7WGR
 
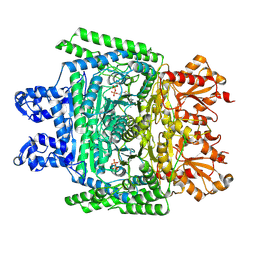 | | Cryo-electron microscopic structure of the 2-oxoglutarate dehydrogenase (E1) component of the human alpha-ketoglutarate (2-oxoglutarate) dehydrogenase complex | | Descriptor: | 2-oxoglutarate dehydrogenase, mitochondrial, CALCIUM ION, ... | | Authors: | Yu, X, Yang, W, Zhong, Y.H, Ma, X.M, Gao, Y.Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for the activity and regulation of human alpha-ketoglutarate dehydrogenase revealed by Cryo-EM
Biochem.Biophys.Res.Commun., 602, 2022
|
|
8JJ8
 
 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist | | Descriptor: | Beta-2 adrenergic receptor,Soluble cytochrome b562, ~{N}-[5-[(1~{R})-2-[[(2~{R})-1-(4-methoxyphenyl)propan-2-yl]amino]-1-oxidanyl-ethyl]-2-oxidanyl-phenyl]methanamide | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJL
 
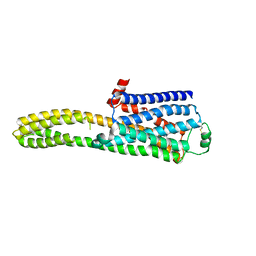 | | cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist | | Descriptor: | Beta-2 adrenergic receptor,Soluble cytochrome b562, Olodaterol | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJO
 
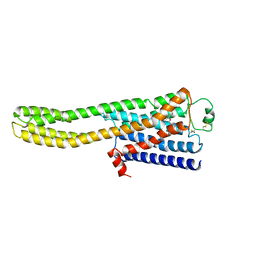 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Beta-2 adrenergic receptor,Soluble cytochrome b562 | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
6WOK
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 6 | | Descriptor: | (1R,3R)-1-(2,6-difluoro-4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Labadie, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Discovery of GNE-149 as a Full Antagonist and Efficient Degrader of Estrogen Receptor alpha for ER+ Breast Cancer.
Acs Med.Chem.Lett., 11, 2020
|
|
6DF6
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 16ab | | Descriptor: | (8R)-8-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-1,8-dihydro-2H-[1]benzopyrano[4,3-d][1]benzoxepine-5,11-diol, Estrogen receptor, GLYCEROL | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Ortwine, D.F, Nettles, K.W, Nwachukwu, J.C. | | Deposit date: | 2018-05-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6DFN
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 16aa | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, (8S)-8-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-1,8-dihydro-2H-[1]benzopyrano[4,3-d][1]benzoxepine-5,11-diol, Estrogen receptor, ... | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Ortwine, D.F, Nettles, K.W, Nwachukwu, J.C. | | Deposit date: | 2018-05-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected equivalent potency of a constrained chromene enantiomeric pair rationalized by co-crystal structures in complex with estrogen receptor alpha.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
4F3I
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Joshua, J, Zhou, M.-M, Plotnikov, A.N. | | Deposit date: | 2012-05-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Down-regulation of NF-{kappa}B Transcriptional Activity in HIV-associated Kidney Disease by BRD4 Inhibition.
J.Biol.Chem., 287, 2012
|
|
7MSA
 
 | | GDC-9545 in complex with estrogen receptor alpha | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, 3-[(1R,3R)-1-(2,6-difluoro-4-{[1-(3-fluoropropyl)azetidin-3-yl]amino}phenyl)-3-methyl-1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl]-2,2-difluoropropan-1-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zbieg, J.R, Wang, X, Ortwine, D.F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-02 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | GDC-9545 (Giredestrant): A Potent and Orally Bioavailable Selective Estrogen Receptor Antagonist and Degrader with an Exceptional Preclinical Profile for ER+ Breast Cancer.
J.Med.Chem., 64, 2021
|
|
6BI6
 
 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
8J7E
 
 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
8EXL
 
 | | Crystal structure of PI3K-alpha in complex with taselisib | | Descriptor: | 2-methyl-2-(4-{2-[3-methyl-1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-1H-pyrazol-1-yl)propanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXO
 
 | | Crystal structure of PI3K-alpha in complex with compound 19 | | Descriptor: | 1-{(4S,11aM)-2-[(4R)-2-oxo-4-(propan-2-yl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-prolinamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXU
 
 | | Crystal structure of PI3K-alpha in complex with compound 30 | | Descriptor: | (2S)-2-cyclopropyl-2-({(4S,11aM)-2-[(4S)-2-oxo-4-(trifluoromethyl)-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}amino)acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
8EXV
 
 | | Crystal structure of PI3K-alpha in complex with compound 32 | | Descriptor: | N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Kiefer, J.R, Eigenbrot, C, Staben, S.T, Hanan, E.J, Wallweber, H.J.A, Ultsch, M, Braun, M.G, Friedman, L.S, Purkey, H.E. | | Deposit date: | 2022-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of GDC-0077 (Inavolisib), a Highly Selective Inhibitor and Degrader of Mutant PI3K alpha.
J.Med.Chem., 65, 2022
|
|
7TDW
 
 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
7TDX
 
 | | Structure of FOXP3-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*AP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*TP*AP*AP*AP*CP*AP*AP*AP*TP*TP*T)-3'), Forkhead box P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2022-01-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The transcription factor FoxP3 can fold into two dimerization states with divergent implications for regulatory T cell function and immune homeostasis.
Immunity, 55, 2022
|
|
5IME
 
 | | Crystal structure of P21-activated kinase 1 (PAK1) in complex with compound 9 | | Descriptor: | 8-(3-aminopropyl)-6-[2-chloro-4-(3-methyl-2-oxopyrazin-1(2H)-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 1 | | Authors: | Li, D, Wang, W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Chemically Diverse Group I p21-Activated Kinase (PAK) Inhibitors Impart Acute Cardiovascular Toxicity with a Narrow Therapeutic Window.
J.Med.Chem., 59, 2016
|
|
