6E5U
 
 | |
8I4S
 
 | | the complex structure of SARS-CoV-2 Mpro with D8 | | Descriptor: | 3-(4-fluoranyl-3-methyl-phenyl)-2-(2-methylpropyl)-5,6,7-tris(oxidanyl)quinazolin-4-one, ORF1a polyprotein | | Authors: | Lu, M. | | Deposit date: | 2023-01-21 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of quinazolin-4-one-based non-covalent inhibitors targeting the severe acute respiratory syndrome coronavirus 2 main protease (SARS-CoV-2 M pro ).
Eur.J.Med.Chem., 257, 2023
|
|
7W16
 
 | | Complex structure of alginate lyase AlyV with M8 | | Descriptor: | GLYCEROL, alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W13
 
 | | Complex structure of alginate lyase PyAly with M8 | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W18
 
 | | Complex structure of alginate lyase PyAly with M5 | | Descriptor: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7T32
 
 | | CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385 | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein | | Authors: | Zhang, K.H, Wu, H, Hoppe, N, Manglik, A, Cheng, Y.F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
7W12
 
 | | Complex structure of alginate lyase AlyB-OU02 with G9 | | Descriptor: | Alginate lyase, SULFATE ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7JSO
 
 | |
3TKY
 
 | | Monolignol o-methyltransferase (momt) | | Descriptor: | (Iso)eugenol O-methyltransferase, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Bhuiya, M.W, Liu, C.J. | | Deposit date: | 2011-08-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | An engineered monolignol 4-o-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis.
Plant Cell, 24, 2012
|
|
6BG9
 
 | |
7WYG
 
 | | Crystal structure of P450BSbeta-L78I/Q85H/G290I variant in complex with palmitic acid. | | Descriptor: | Cytochrome P450 152A1, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Li, F, He, C, Wang, X. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic Enantioselective beta-Hydroxylation of Unactivated C-H Bonds in Aliphatic Carboxylic Acids.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4R7H
 
 | | Crystal structure of FMS KINASE domain with a small molecular inhibitor, PLX3397 | | Descriptor: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Zhang, Y, Zhang, K, Zhang, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor.
N Engl J Med, 373, 2015
|
|
7ONT
 
 | | PARP1 catalytic domain in complex with a selective pyridine carboxamide-based inhibitor (compound 22) | | Descriptor: | 5-[4-[(3-ethyl-2-oxidanylidene-1~{H}-quinolin-7-yl)methyl]piperazin-1-yl]-~{N}-methyl-pyridine-2-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M.J, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONS
 
 | | PARP1 catalytic domain in complex with isoquinolone-based inhibitor (compound 16) | | Descriptor: | 7-[[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]methyl]-3-ethyl-1~{H}-quinolin-2-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Heightman, T.D, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
6ZY3
 
 | | Cryo-EM structure of MlaFEDB in complex with phospholipid | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY2
 
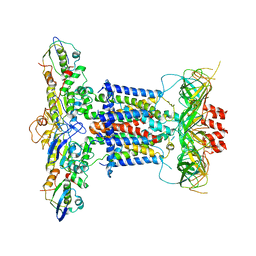 | | Cryo-EM structure of apo MlaFEDB | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, Toluene tolerance protein Ttg2A, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY4
 
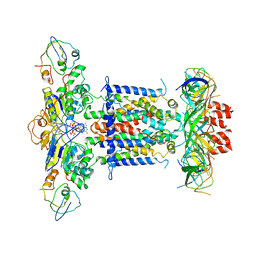 | | Cryo-EM structure of MlaFEDB in complex with ADP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZY9
 
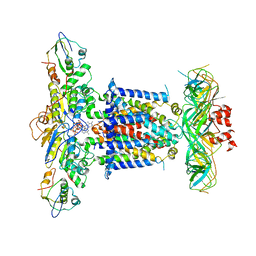 | | Cryo-EM structure of MlaFEDB in complex with AMP-PNP | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, MAGNESIUM ION, ... | | Authors: | Dong, C.J, Dong, H.H. | | Deposit date: | 2020-07-30 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8WCT
 
 | |
4XNW
 
 | | The human P2Y1 receptor in complex with MRS2500 | | Descriptor: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
4XNV
 
 | | The human P2Y1 receptor in complex with BPTU | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2-(2-tert-butylphenoxy)pyridin-3-yl]-3-[4-(trifluoromethoxy)phenyl]urea, CHOLESTEROL, ... | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
6DQB
 
 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ8
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR N49 i.e. 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, 2-[(S)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ6
 
 | |
