6U59
 
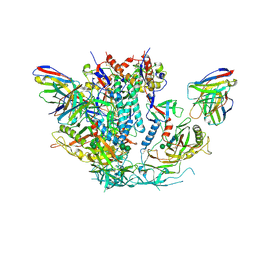 | | HIV-1 B41 SOSIP.664 in complex with rabbit antibody 13B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SOSIP.664 gp120,SOSIP.664 gp120, ... | | Authors: | Yang, Y.R, Ward, A.B. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Autologous Antibody Responses to an HIV Envelope Glycan Hole Are Not Easily Broadened in Rabbits.
J.Virol., 94, 2020
|
|
6NXJ
 
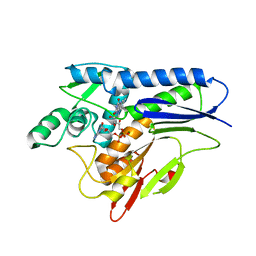 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6NXI
 
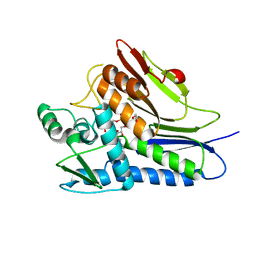 | | Flavin Transferase ApbE from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
8SIT
 
 | |
8SIR
 
 | |
8SIQ
 
 | |
8SIS
 
 | |
8DGV
 
 | |
8DGW
 
 | |
8DGU
 
 | |
8DGX
 
 | |
7KNX
 
 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNW
 
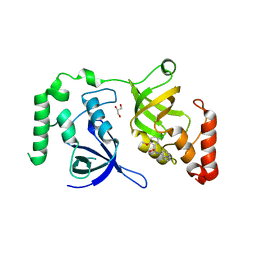 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KSG
 
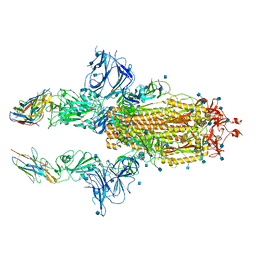 | | SARS-CoV-2 spike in complex with nanobodies E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 glycoprotein, Spike glycoprotein | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-22 | | Release date: | 2021-01-20 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7LM8
 
 | |
7B17
 
 | | SARS-CoV-spike RBD bound to two neutralising nanobodies. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama,SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-10 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B14
 
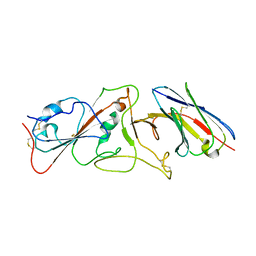 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B18
 
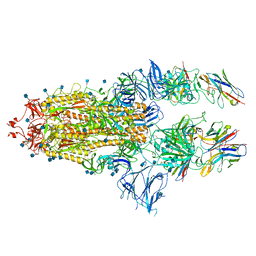 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN4
 
 | |
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7T7B
 
 | |
7E1L
 
 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E1N
 
 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | Descriptor: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7FEW
 
 | |
7FF0
 
 | |
