2IXB
 
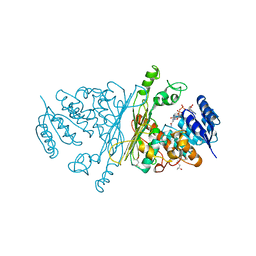 | | Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
5GZR
 
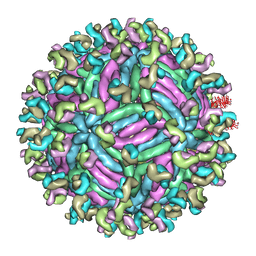 | | Zika virus E protein complexed with a neutralizing antibody Z23-Fab | | Descriptor: | Z23 Fab heavy chain, Z23 Fab light chain, structural protein E, ... | | Authors: | Gao, G.G, Shi, Y, Peng, R, Liu, S. | | Deposit date: | 2016-10-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
5IMX
 
 | |
8HF8
 
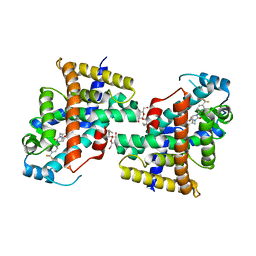 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist V1 | | Descriptor: | 2-[4-[[2,5-bis(oxidanylidene)-3-[4-(trifluoromethyl)phenyl]imidazolidin-1-yl]methyl]-2,6-dimethyl-phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of the First Subnanomolar PPAR alpha / delta Dual Agonist for the Treatment of Cholestatic Liver Diseases.
J.Med.Chem., 66, 2023
|
|
7DL2
 
 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
6M1K
 
 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
5IVL
 
 | | CshA Helicase | | Descriptor: | DEAD-box ATP-dependent RNA helicase CshA, SULFATE ION | | Authors: | Huen, J, Lin, C.-L, Yi, W.-L, Li, C.-L, Yuan, H. | | Deposit date: | 2016-03-21 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into a Unique Dimeric DEAD-Box Helicase CshA that Promotes RNA Decay.
Structure, 25, 2017
|
|
7W0G
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist H11 | | Descriptor: | 2-[2,6-dimethyl-4-[[5-oxidanylidene-4-[4-(trifluoromethyloxy)phenyl]-1,2,4-triazol-1-yl]methyl]phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR alpha / delta Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
4PBP
 
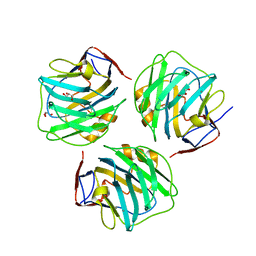 | | crystal structure of zebrafish short-chain pentraxin protein | | Descriptor: | C-reactive protein, CALCIUM ION, GLYCEROL | | Authors: | Chen, R, Qi, J.X, George, F.G, Xia, C. | | Deposit date: | 2014-04-13 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Crystal structures for short-chain pentraxin from zebrafish demonstrate a cyclic trimer with new recognition and effector faces.
J.Struct.Biol., 189, 2015
|
|
4PBO
 
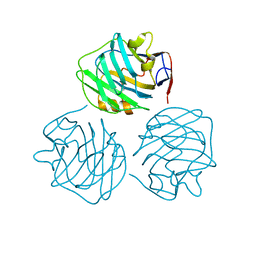 | |
4Q4K
 
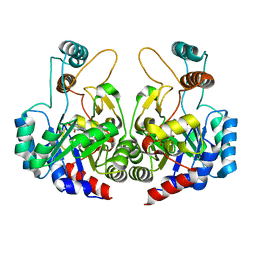 | | Crystal structure of nitronate monooxygenase from Pseudomonas aeruginosa PAO1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Nitronate Monooxygenase | | Authors: | Salvi, F, Agniswamy, J, Gadda, G, Weber, I.T. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Combined Structural and Kinetic Characterization of a Bacterial Nitronate Monooxygenase from Pseudomonas aeruginosa PAO1 Establishes NMO Class I and II.
J.Biol.Chem., 289, 2014
|
|
4QAS
 
 | | 1.27 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAQ
 
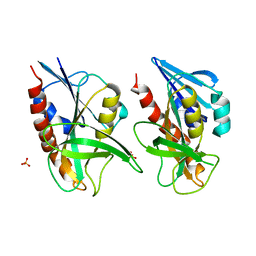 | | 1.58 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QFB
 
 | | 1.99 A resolution structure of SeMet-CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAT
 
 | | 1.75 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis bound to MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAR
 
 | | 1.45 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis bound to Adenine | | Descriptor: | ADENINE, CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
3DBG
 
 | |
3EL3
 
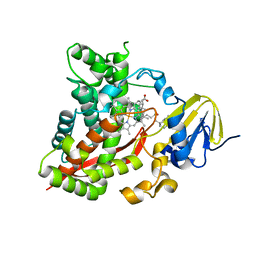 | | Distinct Monooxygenase and Farnesene Synthase Active Sites in Cytochrome P450 170A1 | | Descriptor: | (3S,3aR,6S)-3,7,7,8-tetramethyl-2,3,4,5,6,7-hexahydro-1H-3a,6-methanoazulene, PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Zhao, B, Waterman, M.R. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site
J.Biol.Chem., 284, 2009
|
|
7C6O
 
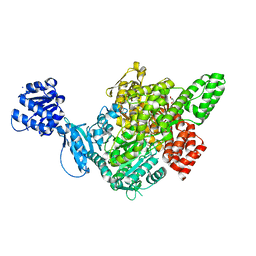 | |
3GV3
 
 | |
3NYE
 
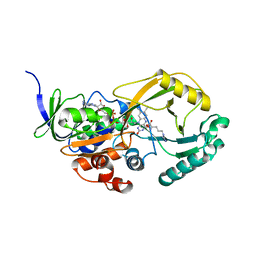 | |
8H56
 
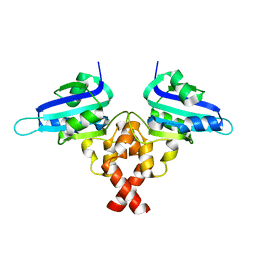 | | Crystal structure of Rep' of porcine circovirus type 2 | | Descriptor: | Isoform Rep' of Replication-associated protein | | Authors: | Guan, S.Y, Song, Y.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of the dimerized of porcine circovirus type II replication-related protein Rep'.
Proteins, 91, 2023
|
|
3SM8
 
 | |
2NZA
 
 | |
2NZ5
 
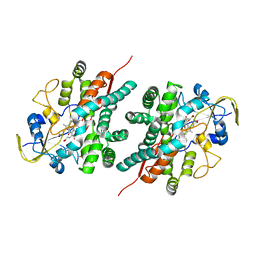 | | Structure and Function Studies of Cytochrome P450 158A1 from Streptomyces coelicolor A3(2) | | Descriptor: | Cytochrome P450 CYP158A1, PROTOPORPHYRIN IX CONTAINING FE, naphthalene-1,2,4,5,7-pentol | | Authors: | Zhao, B, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2006-11-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different binding modes of two flaviolin substrate molecules in cytochrome P450 158A1 (CYP158A1) compared to CYP158A2.
Biochemistry, 46, 2007
|
|
