2G3F
 
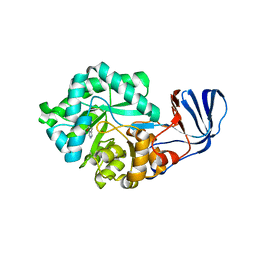 | | Crystal Structure of imidazolonepropionase complexed with imidazole-4-acetic acid sodium salt, a substrate homologue | | Descriptor: | 2H-IMIDAZOL-4-YLACETIC ACID, Imidazolonepropionase, ZINC ION | | Authors: | Yu, Y, Liang, Y.H, Su, X.D. | | Deposit date: | 2006-02-19 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A catalytic mechanism revealed by the crystal structures of the imidazolonepropionase from Bacillus subtilis
J.Biol.Chem., 281, 2006
|
|
4GIF
 
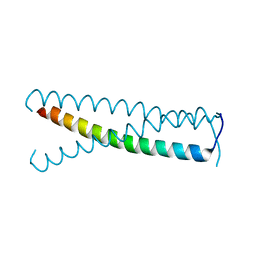 | | C-terminal coiled-coil domain of transient receptor potential channel TRPP3 (PKD2L1, Polycystin-L) | | Descriptor: | Polycystic kidney disease 2-like 1 protein | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Dobbins, S, Zhang, W.K, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2012-08-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of the assembly of an acid-sensing receptor ion channel complex.
Nat Commun, 3, 2012
|
|
1O5T
 
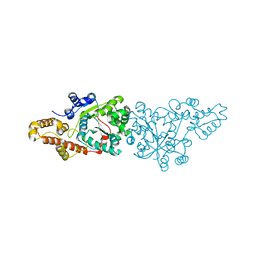 | | Crystal structure of the aminoacylation catalytic fragment of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Yu, Y, Liu, Y, Shen, N, Xu, X, Jia, J, Jin, Y, Arnold, E, Ding, J. | | Deposit date: | 2003-10-05 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Tryptophanyl-tRNA Synthetase Catalytic Fragment
J.BIOL.CHEM., 279, 2004
|
|
7SDE
 
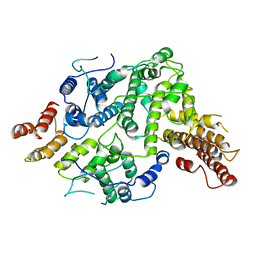 | | Cryo-EM structure of Nse5/6 heterodimer | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-09-29 | | Release date: | 2021-10-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of Nse5/6 complex with the C terminal part of Nse5
To Be Published
|
|
7DOR
 
 | | E. coli GyrB ATPase domain in complex with 4-nitropheno | | Descriptor: | 1H-benzimidazol-2-amine, DNA gyrase subunit B, P-NITROPHENOL, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DPR
 
 | |
7DPS
 
 | |
7DQH
 
 | | E. coli GyrB ATPase domain in complex with 2-hydroxybenzamide | | Descriptor: | 1H-benzimidazol-2-amine, DNA gyrase subunit B, PHOSPHATE ION, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-23 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQI
 
 | | E. coli GyrB ATPase domain in complex with Esculetin | | Descriptor: | 1H-benzimidazol-2-amine, 6,7-bis(oxidanyl)chromen-2-one, DNA gyrase subunit B, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-23 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQJ
 
 | | E. coli GyrB ATPase domain in complex with 3,4-Dihydroxyacetophenone | | Descriptor: | 1-[3,4-bis(oxidanyl)phenyl]ethanone, 1H-benzimidazol-2-amine, DNA gyrase subunit B, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQF
 
 | |
7DQU
 
 | | E. coli GyrB ATPase domain in complex with methyl 4-hydroxybenzoate | | Descriptor: | 1H-benzimidazol-2-amine, 4-HYDROXY-BENZOIC ACID METHYL ESTER, DNA gyrase subunit B, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQL
 
 | | E. coli GyrB ATPase domain in complex with 4-chlorobenzene-1,2-diol | | Descriptor: | 1H-benzimidazol-2-amine, 4-CHLOROBENZENE-1,2-DIOL, DNA gyrase subunit B, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQM
 
 | | E. coli GyrB ATPase domain in complex with Naringenin | | Descriptor: | 1H-benzimidazol-2-amine, DNA gyrase subunit B, PHOSPHATE ION, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQS
 
 | | E. coli GyrB ATPase domain in complex with 2-chlorophenol | | Descriptor: | 1H-benzimidazol-2-amine, 2-CHLOROPHENOL, DNA gyrase subunit B, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
7DQW
 
 | | E. coli GyrB ATPase domain in complex with 4-chlorophenol | | Descriptor: | 1H-benzimidazol-2-amine, 4-chlorophenol, DNA gyrase subunit B, ... | | Authors: | Yu, Y, Zhou, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification of new building blocks by fragment screening for discovering GyrB inhibitors.
Bioorg.Chem., 114, 2021
|
|
1XTQ
 
 | | Structure of small GTPase human Rheb in complex with GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Yu, Y, Ding, J. | | Deposit date: | 2004-10-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Unique Biological Function of Small GTPase RHEB
J.Biol.Chem., 280, 2005
|
|
1XTS
 
 | | Structure of small GTPase human Rheb in complex with GTP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Yu, Y, Ding, J. | | Deposit date: | 2004-10-24 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Unique Biological Function of Small GTPase RHEB
J.Biol.Chem., 280, 2005
|
|
1XTR
 
 | |
3IPM
 
 | |
4HK4
 
 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
4HPW
 
 | | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine | | Descriptor: | 3-methoxy-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine
To be Published
|
|
7KEO
 
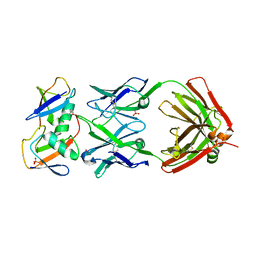 | | Crystal structure of K29-linked di-ubiquitin in complex with synthetic antigen binding fragment | | Descriptor: | PHOSPHATE ION, Synthetic antigen binding fragment, heavy chain, ... | | Authors: | Yu, Y, Zheng, Q, Erramilli, S, Pan, M, Kossiakoff, A, Liu, L, Zhao, M. | | Deposit date: | 2020-10-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Nat.Chem.Biol., 17, 2021
|
|
7TVE
 
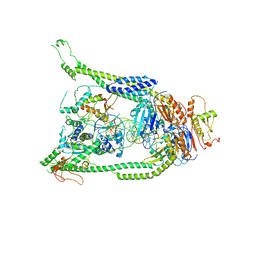 | | ATP and DNA bound SMC5/6 core complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (68-MER), DNA (78-MER), ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2022-02-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of DNA-bound Smc5/6 reveals DNA clamping enabled by multi-subunit conformational changes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4M5C
 
 | | Crystal Structure of an Truncated Transition metal Transporter | | Descriptor: | COBALT (II) ION, Cobalamin biosynthesis protein CbiM, HEXANE-1,6-DIOL | | Authors: | Yu, Y, Zhou, M.Z, Gu, J.K. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
