3C1S
 
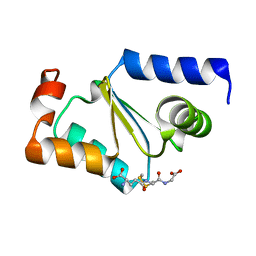 | | Crystal structure of GRX1 in glutathionylated form | | Descriptor: | GLUTATHIONE, Glutaredoxin-1 | | Authors: | Yu, J, Zhou, C.Z. | | Deposit date: | 2008-01-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathionylation-triggered conformational changes of glutaredoxin Grx1 from the yeast Saccharomyces cerevisiae.
Proteins, 72, 2008
|
|
6W25
 
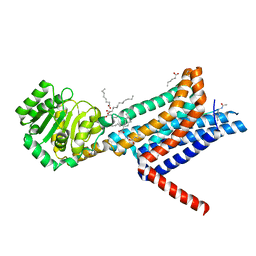 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
3C1R
 
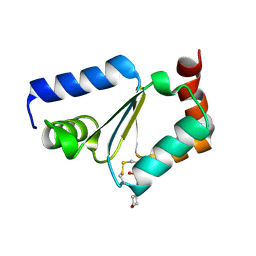 | | Crystal structure of oxidized GRX1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutaredoxin-1 | | Authors: | Yu, J, Zhou, C.Z. | | Deposit date: | 2008-01-24 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glutathionylation-triggered conformational changes of glutaredoxin Grx1 from the yeast Saccharomyces cerevisiae.
Proteins, 72, 2008
|
|
8QOT
 
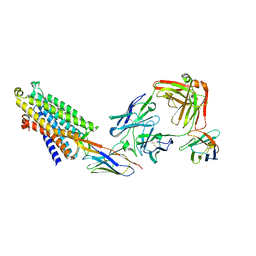 | | Structure of the mu opioid receptor bound to the antagonist nanobody NbE | | Descriptor: | Anti-Fab Nanobody, Mu-type opioid receptor, NabFab HC, ... | | Authors: | Yu, J, Kumar, A, Zhang, X, Martin, C, Raia, P, Manglik, A, Ballet, S, Boland, A, Stoeber, M. | | Deposit date: | 2023-09-29 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of mu-Opioid Receptor-Targeting by a Nanobody Antagonist.
Biorxiv, 2023
|
|
2QJL
 
 | | Crystal structure of Urm1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin-related modifier 1 | | Authors: | Yu, J, Zhou, C.Z. | | Deposit date: | 2007-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of the dimeric Urm1 from the yeast Saccharomyces cerevisiae.
Proteins, 71, 2008
|
|
7LDE
 
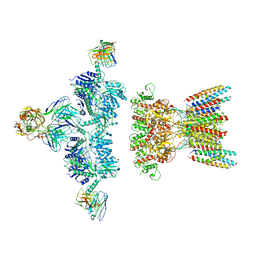 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LEP
 
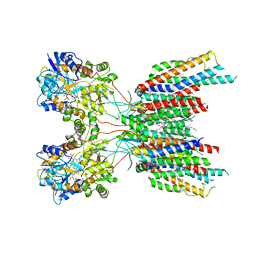 | | The composite LBD-TMD structure combined from all hippocampal AMPAR subtypes at 3.25 Angstrom resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol, DECANE, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-14 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
7LDD
 
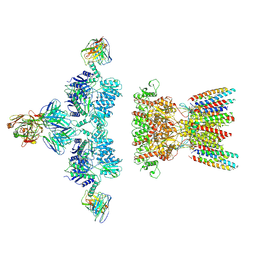 | | native AMPA receptor | | Descriptor: | 11B8 scFv, 15F1 Fab heavy chain, 15F1 Fab light chain, ... | | Authors: | Yu, J, Rao, P, Gouaux, E. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hippocampal AMPA receptor assemblies and mechanism of allosteric inhibition.
Nature, 594, 2021
|
|
3W7A
 
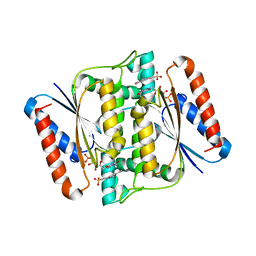 | | Crystal Structure of azoreductase AzrC fin complex with sulfone-modified azo dye Acid Red 88 | | Descriptor: | 4-[(E)-(2-hydroxynaphthalen-1-yl)diazenyl]naphthalene-1-sulfonic acid, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yu, J, Ogata, D, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3W78
 
 | | Crystal Structure of azoreductase AzrC in complex with NAD(P)-inhibitor Cibacron Blue | | Descriptor: | CIBACRON BLUE, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Yu, J, Ogata, D, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2HQM
 
 | |
5ZT0
 
 | |
5ZQV
 
 | |
7YGG
 
 | |
4EGX
 
 | | Crystal structure of KIF1A CC1-FHA tandem | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Kinesin-like protein KIF1A | | Authors: | Yu, J, Huo, L, Yue, Y, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
3SHU
 
 | | Crystal structure of ZO-1 PDZ3 | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
3SHW
 
 | | Crystal structure of ZO-1 PDZ3-SH3-Guk supramodule complex with Connexin-45 peptide | | Descriptor: | Gap junction gamma-1 protein, Tight junction protein ZO-1 | | Authors: | Yu, J, Pan, L, Chen, J, Yu, H, Zhang, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of the PDZ3-SH3-GuK Tandem of ZO-1 Suggests a Supramodular Organization of the Conserved MAGUK Family Scaffold Core
To be Published
|
|
4YMX
 
 | |
4YMV
 
 | | Crystal structure of an amino acid ABC transporter with ATPs | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMS
 
 | | Crystal structure of an amino acid ABC transporter | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMU
 
 | | Crystal structure of an amino acid ABC transporter complex with arginines and ATPs | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMW
 
 | | Crystal structure of an amino acid ABC transporter with histidines | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YMT
 
 | | Crystal structure of an amino acid ABC transporter complex with arginines | | Descriptor: | ABC-type amino acid transport system, permease component, ABC-type polar amino acid transport system, ... | | Authors: | Ge, J, Yu, J, Yang, M. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural basis for substrate specificity of an amino acid ABC transporter
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3TFM
 
 | | Myosin X PH1N-PH2-PH1C tandem | | Descriptor: | Myosin X, PHOSPHATE ION | | Authors: | Yu, J, Lu, Q, Yan, J, Wei, Z, Zhang, M. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of the myosin X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor
MOLECULAR BIOLOGY OF THE CELL, 22, 2011
|
|
3HPM
 
 | |
