5IC9
 
 | | Structure of the CTD complex of Utp12 and Utp13 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5IC8
 
 | | Structure of UTP6 | | Descriptor: | Putative uncharacterized protein | | Authors: | Zhang, C, Ye, K. | | Deposit date: | 2016-02-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits
Nucleic Acids Res., 44, 2016
|
|
5YAX
 
 | | Crystal structure of a human neutralizing antibody bound to a HBV preS1 peptide | | Descriptor: | Large envelope protein, SODIUM ION, scFv1 antibody | | Authors: | Liu, X, Zheng, S, Ye, K, Sui, J. | | Deposit date: | 2017-09-02 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A potent human neutralizing antibody Fc-dependently reduces established HBV infections
Elife, 6, 2017
|
|
4QMF
 
 | | Structure of the Krr1 and Faf1 complex from Saccharomyces cerevisiae | | Descriptor: | KRR1 small subunit processome component, Protein FAF1 | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Interaction between ribosome assembly factors Krr1 and Faf1 is essential for formation of small ribosomal subunit in yeast
J.Biol.Chem., 289, 2014
|
|
4J0X
 
 | | Structure of Rrp9 | | Descriptor: | Ribosomal RNA-processing protein 9 | | Authors: | Zhang, L, Lin, J, Ye, K. | | Deposit date: | 2013-01-31 | | Release date: | 2013-06-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural and functional analysis of the U3 snoRNA binding protein Rrp9.
Rna, 19, 2013
|
|
4J0W
 
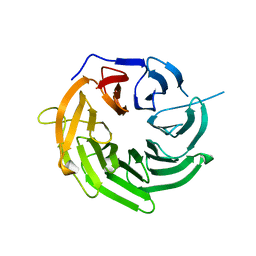 | | Structure of U3-55K | | Descriptor: | U3 small nucleolar RNA-interacting protein 2 | | Authors: | Zhang, L, Lin, J, Ye, K. | | Deposit date: | 2013-01-31 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional analysis of the U3 snoRNA binding protein Rrp9.
Rna, 19, 2013
|
|
4RKG
 
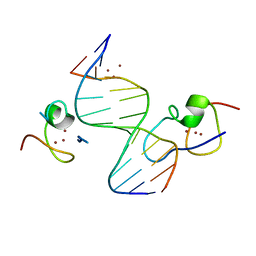 | | Structure of the MSL2 CXC domain bound with a non-specific (GC)6 DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*C)-3'), E3 ubiquitin-protein ligase msl-2, ZINC ION | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
4RKH
 
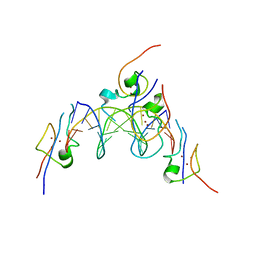 | | Structure of the MSL2 CXC domain bound with a specific MRE sequence | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*CP*TP*CP*GP*CP*TP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*GP*AP*GP*CP*GP*AP*GP*AP*TP*GP*GP*AP*T)-3'), E3 ubiquitin-protein ligase msl-2, ... | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of X chromosome DNA recognition by the MSL2 CXC domain during Drosophila dosage compensation.
Genes Dev., 28, 2014
|
|
4S3H
 
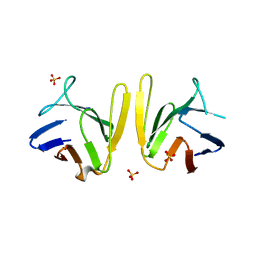 | | Crystal structure of S. pombe Mdb1 FHA domain | | Descriptor: | Mdb1, SULFATE ION | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Dimerization Mediated by a Divergent Forkhead-associated Domain Is Essential for the DNA Damage and Spindle Functions of Fission Yeast Mdb1.
J.Biol.Chem., 290, 2015
|
|
2LUA
 
 | | Solution structure of CXC domain of MSL2 | | Descriptor: | Protein male-specific lethal-2, ZINC ION | | Authors: | Feng, Y, Ye, K, Zheng, S, Wang, J. | | Deposit date: | 2012-06-09 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MSL2 CXC Domain Reveals an Unusual Zn(3)Cys(9) Cluster and Similarity to Pre-SET Domains of Histone Lysine Methyltransferases.
Plos One, 7, 2012
|
|
2MBY
 
 | | NMR Structure of Rrp7 C-terminal Domain | | Descriptor: | Ribosomal RNA-processing protein 7 | | Authors: | Lin, J, Feng, Y, Ye, K. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An RNA-Binding Complex Involved in Ribosome Biogenesis Contains a Protein with Homology to tRNA CCA-Adding Enzyme.
Plos Biol., 11, 2013
|
|
6IF4
 
 | | Crystal structure of Tbtudor | | Descriptor: | Histone acetyltransferase | | Authors: | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | Deposit date: | 2018-09-18 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.934 Å) | | Cite: | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
1I5T
 
 | | SOLUTION STRUCTURE OF CYANOFERRICYTOCHROME C | | Descriptor: | CYANIDE ION, CYTOCHROME C, HEME C | | Authors: | Yao, Y, Qian, C, Ye, K, Wang, J, Tang, W. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cyanoferricytochrome c: ligand-controlled conformational flexibility and electronic structure of the heme moiety.
J.Biol.Inorg.Chem., 7, 2002
|
|
4ZLK
 
 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
5YDU
 
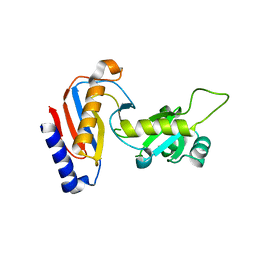 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
5GIP
 
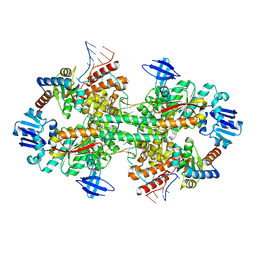 | | Crystal structure of box C/D RNP with 13 nt guide regions and 11 nt substrates | | Descriptor: | 50S ribosomal protein L7Ae, C/D RNA, C/D box methylation guide ribonucleoprotein complex aNOP56 subunit, ... | | Authors: | Yang, Z, Lin, J, Ye, K. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.129 Å) | | Cite: | Box C/D guide RNAs recognize a maximum of 10 nt of substrates
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5YMA
 
 | | Crystal structure of ribosome assembly factor Efg1 | | Descriptor: | Putative rRNA processing protein | | Authors: | Shu, S, Ye, K. | | Deposit date: | 2017-10-21 | | Release date: | 2018-01-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Structural and functional analysis of ribosome assembly factor Efg1.
Nucleic Acids Res., 46, 2018
|
|
5YZ4
 
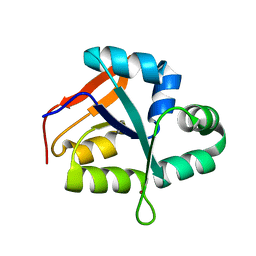 | | Structure of the PIN domain endonuclease Utp24 | | Descriptor: | CALCIUM ION, ZINC ION, rRNA-processing protein fcf1 | | Authors: | Du, Y, An, W, Ye, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Structural and functional analysis of Utp24, an endonuclease for processing 18S ribosomal RNA.
Plos One, 13, 2018
|
|
5GIO
 
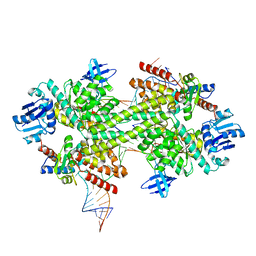 | | Crystal structure of box C/D RNP with 12 nt guide regions and 13 nt substrates | | Descriptor: | 50S ribosomal protein L7Ae, C/D RNA, C/D box methylation guide ribonucleoprotein complex aNOP56 subunit, ... | | Authors: | Yang, Z, Lin, J, Ye, K. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.604 Å) | | Cite: | Box C/D guide RNAs recognize a maximum of 10 nt of substrates
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5Z1G
 
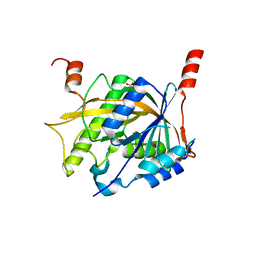 | | Structure of the Brx1 and Ebp2 complex | | Descriptor: | Ribosome biogenesis protein BRX1, SULFATE ION, rRNA-processing protein EBP2 | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2017-12-26 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate
Protein Cell, 10, 2019
|
|
5GIN
 
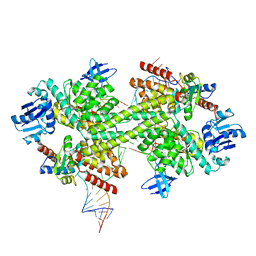 | | Crystal structure of box C/D RNP with 12 nt guide regions and 9 nt substrates | | Descriptor: | 50S ribosomal protein L7Ae, C/D RNA, C/D box methylation guide ribonucleoprotein complex aNOP56 subunit, ... | | Authors: | Yang, Z, Lin, J, Ye, K. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.308 Å) | | Cite: | Box C/D guide RNAs recognize a maximum of 10 nt of substrates
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7XPL
 
 | |
5WYL
 
 | |
5WY4
 
 | |
5WY3
 
 | |
