6NIV
 
 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
7CMZ
 
 | | Crystal Structure of BRCT7/8 in Complex with the APS Motif of PHF8 | | Descriptor: | DNA topoisomerase 2-binding protein 1, Histone lysine demethylase PHF8, POTASSIUM ION, ... | | Authors: | Che, S.Y, Ma, S, Cao, C, Yao, Z, Shi, L, Yang, N. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability.
Sci Adv, 7, 2021
|
|
6MPL
 
 | | Racemic M2-TM I39A crystallized from racemic detergent | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MPN
 
 | | Racemic M2-TM I42E crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
6MPM
 
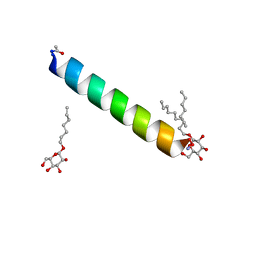 | | Racemic M2-TM I42A crystallized from racemic detergent | | Descriptor: | Matrix protein 2, octyl beta-D-glucopyranoside | | Authors: | Kreitler, D.F, Yao, Z, Mortenson, D.E, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-10-07 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Hendecad Motif Is Preferred for Heterochiral Coiled-Coil Formation.
J. Am. Chem. Soc., 141, 2019
|
|
8JB5
 
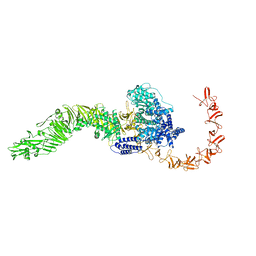 | |
8X2I
 
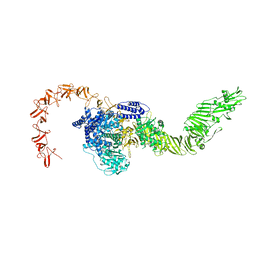 | |
8X2H
 
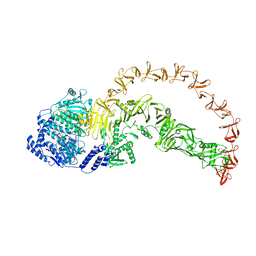 | |
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PER
 
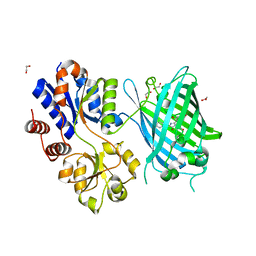 | | Crystal Structure of Ligand-Free iSeroSnFR | | Descriptor: | 1,2-ETHANEDIOL, iSeroSnFR, a soluble, ... | | Authors: | Hartanto, S, Tian, L, Fisher, A.J. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed Evolution of a Selective and Sensitive Serotonin Sensor via Machine Learning.
Cell, 183, 2020
|
|
8XR5
 
 | | Crystal structure of PD-L1 complexed with small molecule inhibitor X18 | | Descriptor: | (2~{R})-2-[[2-(2,1,3-benzoxadiazol-5-ylmethoxy)-5-chloranyl-4-[[2-fluoranyl-3-[3-[3-(4-oxidanylpiperidin-1-yl)propoxy]phenyl]phenyl]methoxy]phenyl]methylamino]-3-oxidanyl-propanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Liu, L. | | Deposit date: | 2024-01-06 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Novel PD-L1 Small-Molecular Inhibitors with Potent In Vivo Anti-tumor Immune Activity.
J.Med.Chem., 67, 2024
|
|
8JP2
 
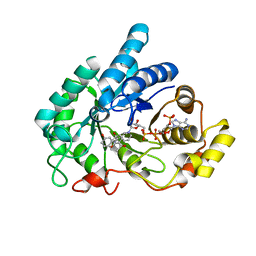 | | Crystal structure of AKR1C1 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
8JP1
 
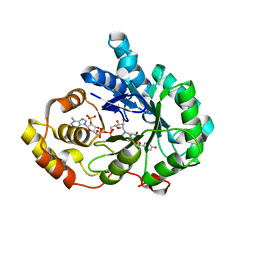 | | Crystal structure of AKR1C3 in complex with DFV | | Descriptor: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X.H, Liu, H, Yao, Z.Q, Zhang, L.P. | | Deposit date: | 2023-06-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of AKR1Cs by liquiritigenin and the structural basis.
Chem.Biol.Interact., 385, 2023
|
|
4Y5U
 
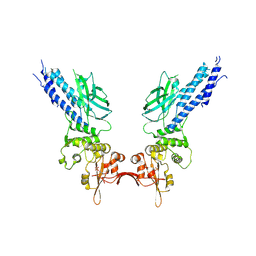 | | Transcription factor | | Descriptor: | NICKEL (II) ION, Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4Y5W
 
 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*GP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*CP*TP*TP*CP*CP*TP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-02-12 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D39
 
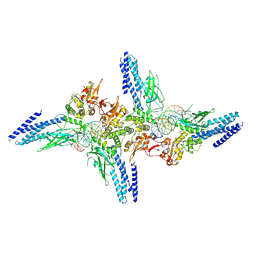 | | Transcription factor-DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*GP*GP*AP*TP*TP*TP*CP*CP*TP*GP*GP*AP*AP*GP*AP*CP*AP*GP*A)-3'), DNA (5'-D(P*TP*CP*TP*GP*TP*CP*TP*TP*CP*CP*AP*GP*GP*AP*AP*AP*TP*CP*CP*AP*T)-3'), Signal transducer and activator of transcription 6 | | Authors: | Li, J, Niu, F, Ouyang, S, Liu, Z. | | Deposit date: | 2015-08-06 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for DNA recognition by STAT6
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5I7U
 
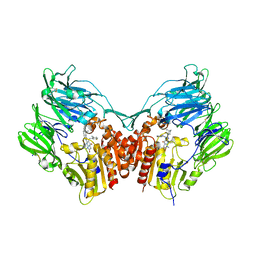 | | Human DPP4 in complex with a novel tricyclic hetero-cycle inhibitor | | Descriptor: | 2-({2-[(3R)-3-aminopiperidin-1-yl]-5-methyl-6,9-dioxo-5,6,7,9-tetrahydro-1H-imidazo[1,2-a]purin-1-yl}methyl)-4-fluorobenzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Novel Tricyclic Heterocycles as Potent and Selective DPP-4 Inhibitors for the Treatment of Type 2 Diabetes.
Acs Med.Chem.Lett., 7, 2016
|
|
5T4B
 
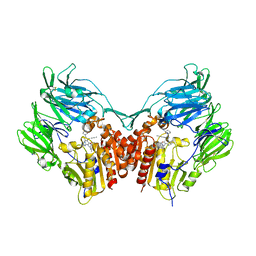 | | Human DPP4 in complex with a ligand 34a | | Descriptor: | 2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-5-[(4-methylquinazolin-2-yl)methyl]-3H-imidazo[2,1-b]purin-4(5H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5T4H
 
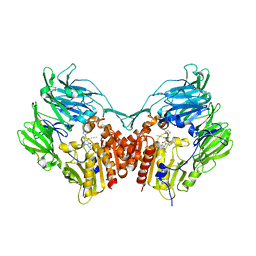 | | Human DPP4 in complex with ligand 34n | | Descriptor: | 2-({2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-4-oxo-3,4-dihydro-5H-imidazo[2,1-b]purin-5-yl}methyl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5T4E
 
 | | Human DPP4 in complex with ligand 19a | | Descriptor: | 2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-6-[(4-methylquinazolin-2-yl)methyl]-6,7,8,9-tetrahydropyrimido[2,1-b]purin-4(3H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5T4F
 
 | | Human DPP4 in complex with ligand 34p | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-({2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-4-oxo-3,4-dihydro-5H-imidazo[2,1-b]purin-5-yl}methyl)benzonitrile, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5IP0
 
 | | PHA Binding Protein PhaP (Phasin) | | Descriptor: | CADMIUM ION, PHA granule-associated protein | | Authors: | Chen, G.Q, Wang, X.Q, Zhao, H.Y. | | Deposit date: | 2016-03-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights on PHA Binding Protein PhaP from Aeromonas hydrophila
Sci Rep, 6, 2016
|
|
9HE1
 
 | | Structure of the Xenorceptide A2-bound E. coli BAM complex (BamABCDE) | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Jakob, R.P, Modaresi, S.M, Maier, T, Hiller, S. | | Deposit date: | 2024-11-13 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Antibiotics that Kill Gram-negative Bacteria by Restructuring the Outer Membrane Protein BamA
Biorxiv, 2024
|
|
7ELG
 
 | | LC3B modificated with a covalent probe | | Descriptor: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Fan, S, Wan, W. | | Deposit date: | 2021-04-10 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5Z5T
 
 | | The first bromodomain of BRD4 with compound BDF-2141 | | Descriptor: | 2-amino-4-(1H-imidazol-1-yl)quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2018-01-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Development and evaluation of a novel series of Nitroxoline-derived BET inhibitors with antitumor activity in renal cell carcinoma.
Oncogenesis, 7, 2018
|
|
