8HE1
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5CTY
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5D7D
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5D7C
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D7R
 
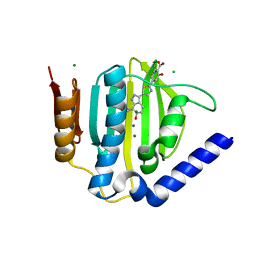 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-hydroxy-5-[5-(6-hydroxy-7-propyl-2H-indazol-3-yl)-1,3-thiazol-2-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-14 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
5VFP
 
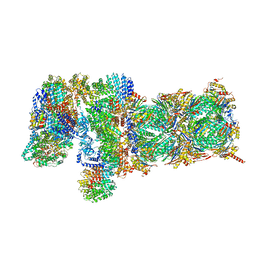 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VHR
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHN
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHJ
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VFR
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5VHQ
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VHO
 
 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 10, 26S proteasome non-ATPase regulatory subunit 2, 26S proteasome regulatory subunit 10B, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
5VFS
 
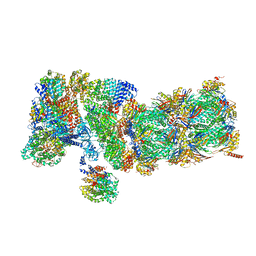 | | Nucleotide-Driven Triple-State Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
5CTW
 
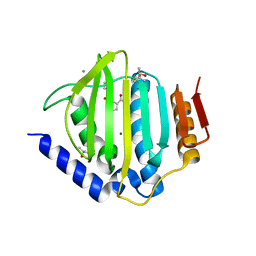 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(butanoylamino)thiophene-3-carboxamide, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Hadfield, A.T, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CTX
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPH
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (3E)-3-(pyridin-3-ylmethylidene)-1,3-dihydro-2H-indol-2-one, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-13 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
1J9I
 
 | | STRUCTURE OF THE DNA BINDING DOMAIN OF THE GPNU1 SUBUNIT OF LAMBDA TERMINASE | | Descriptor: | TERMINASE SMALL SUBUNIT | | Authors: | De Beer, T, Meyer, J, Ortega, M, Yang, Q, Maes, L, Duffy, C, Berton, N, Sippy, J, Overduin, M, Feiss, M, Catalano, C. | | Deposit date: | 2001-05-25 | | Release date: | 2002-08-14 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Insights into specific DNA recognition during the assembly of a viral genome packaging machine.
Mol.Cell, 9, 2002
|
|
3NSN
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin | | Descriptor: | 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3NSM
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
