1PCK
 
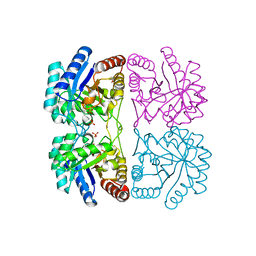 | | Aquifex aeolicus KDO8PS in complex with Z-methyl-PEP | | Descriptor: | 2-(PHOSPHONOOXY)BUTANOIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Wang, J, Xu, X, Grison, C, Petek, S, Coutrot, P, Birck, M, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2003-05-16 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Novel Inhibitors of 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase.
DRUG DES.DISCOVERY, 18, 2003
|
|
3CQV
 
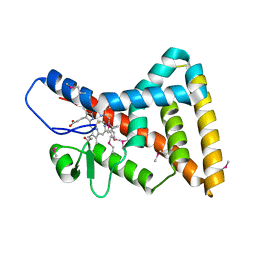 | | Crystal structure of Reverb beta in complex with heme | | Descriptor: | Nuclear receptor subfamily 1 group D member 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, X, Dong, A, Pardee, K.I, Reinking, J, Krause, H, Schuetz, A, Zhang, R, Cui, H, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Savchenko, A, Botchkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta.
Plos Biol., 7, 2009
|
|
3U74
 
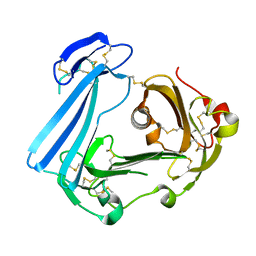 | | Crystal structure of stabilized human uPAR mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the urokinase receptor in a ligand-free form.
J.Mol.Biol., 416, 2012
|
|
3U3L
 
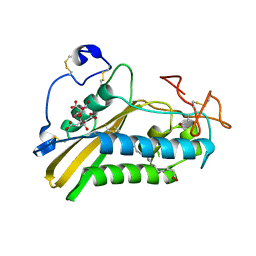 | | Crystal structure of the selenomethionine derivative of tablysin-15 | | Descriptor: | CITRIC ACID, PALMITIC ACID, PRASEODYMIUM ION, ... | | Authors: | Andersen, J.F, Xu, X, Ribeiro, J.M. | | Deposit date: | 2011-10-06 | | Release date: | 2012-02-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain.
J.Biol.Chem., 287, 2012
|
|
3U73
 
 | | Crystal structure of stabilized human uPAR mutant in complex with ATF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor, Urokinase-type plasminogen activator, ... | | Authors: | Huang, M.D, Xu, X, Yuan, C. | | Deposit date: | 2011-10-13 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of the urokinase receptor in a ligand-free form.
J.Mol.Biol., 416, 2012
|
|
5WA5
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor XMD11-50 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Xu, X, Blacklow, S.C. | | Deposit date: | 2017-06-24 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.172 Å) | | Cite: | Structural and Atropisomeric Factors Governing the Selectivity of Pyrimido-benzodiazipinones as Inhibitors of Kinases and Bromodomains.
ACS Chem. Biol., 13, 2018
|
|
8K9E
 
 | |
8K9F
 
 | |
5I6V
 
 | |
2N8I
 
 | | Solution NMR Structure of Designed Protein DA05, Northeast Structural Genomics Consortium (NESG) Target OR626 | | Descriptor: | Designed Protein DA05 | | Authors: | Xu, X, Eletsky, A, Federizon, J.F, Jacobs, T.M, Kuhlman, B, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-10-15 | | Release date: | 2016-01-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design of structurally distinct proteins using strategies inspired by evolution.
Science, 352, 2016
|
|
8HP5
 
 | | Crystal structure of (S)-2-haloacid dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP7
 
 | | Crystal structure of (S)-2-haloacid dehalogenase K152A mutant trapped with (2R)-4-amino-2-hydroxybutanoic acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
8HP6
 
 | | Crystal structure of (S)-2-haloacid dehalogenase D12A mutant | | Descriptor: | (S)-2-haloacid dehalogenase, SODIUM ION | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
7EL9
 
 | | Structure of Machupo virus L polymerase in complex with Z protein and 3'-vRNA (dimeric complex) | | Descriptor: | MANGANESE (II) ION, Machupo virus 3'-vRNA promoter, RING finger protein Z, ... | | Authors: | Peng, R, Xu, X, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
7ELB
 
 | | Structure of Machupo virus L polymerase in complex with Z protein (dimeric form) | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
8HJU
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8HJV
 
 | | Cryo-EM structure of carotenoid-depleted RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8J5O
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 100lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
8J5P
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 2,000lux | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | Authors: | Xu, X, Xin, J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
3Q6T
 
 | | Salivary protein from Lutzomyia longipalpis, Ligand free | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
3Q6P
 
 | | Salivary protein from Lutzomyia longipalpis. Selenomethionine derivative | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
3Q6K
 
 | | Salivary protein from Lutzomyia longipalpis | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID, SEROTONIN | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-02 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
4GE1
 
 | | Structure of the tryptamine complex of the amine binding protein of Rhodnius prolixus | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, Biogenic amine-binding protein, GLYCEROL | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GET
 
 | | Crystal structure of biogenic amine binding protein from Rhodnius prolixus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Biogenic amine-binding protein | | Authors: | Andersen, J.F, Chang, B.W, Xu, X, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5UX6
 
 | | Structure of Human POFUT1 in its apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
