8XOO
 
 | |
8XOP
 
 | |
8XON
 
 | |
4HBN
 
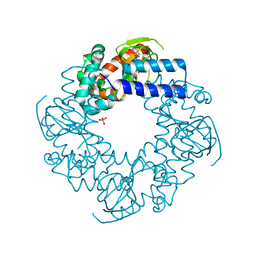 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
7F18
 
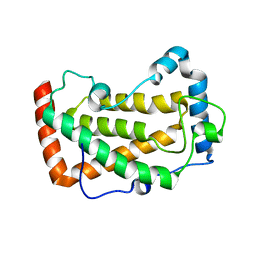 | | Crystal Structure of a mutant of acid phosphatase from Pseudomonas aeruginosa (Q57H/W58P/D135R) | | Descriptor: | Acid phosphatase | | Authors: | Xu, X, Hou, X.D, Song, W, Yin, D.J, Rao, Y.J, Liu, L.M. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
7F17
 
 | | Crystal Structure of acid phosphatase | | Descriptor: | Acid phosphatase | | Authors: | Xu, X, Hou, X.D, Song, W, Rao, Y.J, Liu, L.M, Wu, J. | | Deposit date: | 2021-06-08 | | Release date: | 2021-10-27 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Local Electric Field Modulated Reactivity of Pseudomonas aeruginosa Acid Phosphatase for Enhancing Phosphorylation of l-Ascorbic Acid
Acs Catalysis, 11, 2021
|
|
2JQR
 
 | | Solution model of crosslinked complex of cytochrome c and adrenodoxin | | Descriptor: | Adrenodoxin, mitochondrial, Cytochrome c iso-1, ... | | Authors: | Xu, X, Reinle, W, Hannemann, F, Konarev, P.V, Svergun, D.I, Bernhardt, R, Ubbink, M. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy
J.Am.Chem.Soc., 130, 2008
|
|
2KAJ
 
 | | NMR structure of gallium substituted ferredoxin | | Descriptor: | Ferredoxin-1, GALLIUM (III) ION | | Authors: | Xu, X, Ubbink, M, Knaff, D.B. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the ga-substituted ferredoxin from Synechocystis sp. PCC6803, a mimic of the native protein.
Biochemistry, 49, 2010
|
|
2LY9
 
 | | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F | | Descriptor: | Zinc fingers and homeoboxes protein 1 | | Authors: | Xu, X, Eletsky, A, Mills, J.L, Pulavarti, S.V.S.R.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F
To be Published
|
|
2MIQ
 
 | | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 of Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J | | Descriptor: | Lysine-specific demethylase lid, ZINC ION | | Authors: | Xu, X, Eletsky, A, Shastry, R, Maglaqui, M, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 from Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J
To be Published
|
|
2MIO
 
 | | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A | | Descriptor: | Rho GTPase-activating protein 10 | | Authors: | Xu, X, Eletsky, A, Pulavarti, S.V.S.R.K, Wang, H, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A
To be Published
|
|
2M9W
 
 | | Solution NMR Structure of Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B | | Descriptor: | Transcription factor GATA-4, ZINC ION | | Authors: | Xu, X, Eletsky, A, Lee, D, Kohn, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Transcription Factor GATA-4 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR4783B
To be Published
|
|
2M0C
 
 | | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C | | Descriptor: | Homeobox protein aristaless-like 4 | | Authors: | Xu, X, Eletsky, A, Pulavarti, S, Lee, D, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-10-24 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox Domain of Human ALX4, Northeast Structural Genomics Consortium (NESG) Target HR4490C
To be Published
|
|
2M9X
 
 | | Solution NMR Structure of Microtubule-associated serine/threonine-protein kinase 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9151A | | Descriptor: | Microtubule-associated serine/threonine-protein kinase 1 | | Authors: | Xu, X, Eletsky, A, Shastry, R, Lee, D, Hamilton, K, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Microtubule-associated serine/threonine-protein kinase 1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9151A
To be Published
|
|
2MR6
 
 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR462 | | Descriptor: | De novo designed Protein OR462 | | Authors: | Xu, X, Nivon, L, Federizon, J.F, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR462
To be Published
|
|
5DAN
 
 | |
2PV2
 
 | |
2PV1
 
 | |
2PV3
 
 | |
7CKL
 
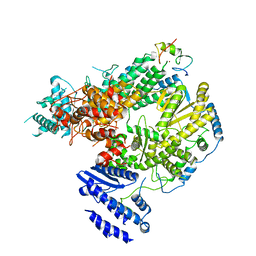 | | Structure of Lassa virus polymerase bound to Z matrix protein | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
7CKM
 
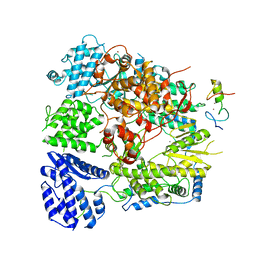 | | Structure of Machupo virus polymerase bound to Z matrix protein (monomeric complex) | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals
Nat Microbiol, 6, 2021
|
|
7ELC
 
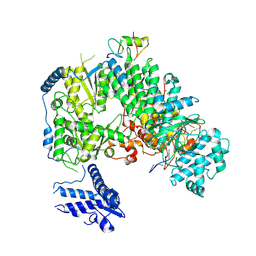 | | Structure of monomeric complex of MACV L bound to Z and 3'-vRNA | | Descriptor: | 3'-vRNA promoter, MANGANESE (II) ION, RING finger protein Z, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
7ELA
 
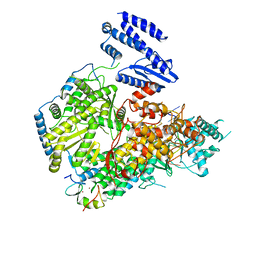 | | Structure of Lassa virus polymerase in complex with 3'-vRNA and Z mutant (F36A) | | Descriptor: | 3-'vRNA promoter, MANGANESE (II) ION, RING finger protein Z, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
4L87
 
 | |
3OTF
 
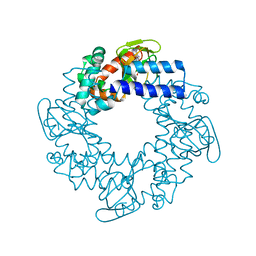 | | Structural basis for the cAMP-dependent gating in human HCN4 channel | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Vysotskaya, Z.V, Liu, Q, Zhou, L. | | Deposit date: | 2010-09-11 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the cAMP-dependent gating in the human HCN4 channel.
J.Biol.Chem., 285, 2010
|
|
