6XN8
 
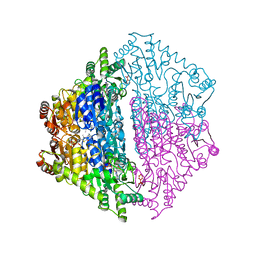 | | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017 | | Descriptor: | 2-hydroxyacyl-CoA lyase 1, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Miller, M.D, Xu, W, Olmos Jr, J.L, Chou, A, Clomburg, J.M, Gonzalez, R, Philips Jr, G.N. | | Deposit date: | 2020-07-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017
To Be Published
|
|
6XOD
 
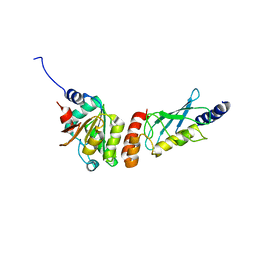 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
3FGA
 
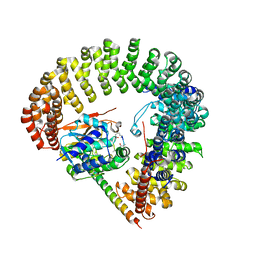 | | Structural Basis of PP2A and Sgo interaction | | Descriptor: | MANGANESE (II) ION, MICROCYSTIN-LR, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Xu, Z, Xu, W. | | Deposit date: | 2008-12-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the PP2A-shugoshin interaction
Mol.Cell, 35, 2009
|
|
6PAX
 
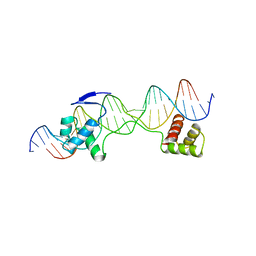 | | CRYSTAL STRUCTURE OF THE HUMAN PAX-6 PAIRED DOMAIN-DNA COMPLEX REVEALS A GENERAL MODEL FOR PAX PROTEIN-DNA INTERACTIONS | | Descriptor: | 26 NUCLEOTIDE DNA, HOMEOBOX PROTEIN PAX-6 | | Authors: | Xu, H.E, Rould, M.A, Xu, W, Epstein, J.A, Maas, R.L, Pabo, C.O. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding.
Genes Dev., 13, 1999
|
|
2Z6H
 
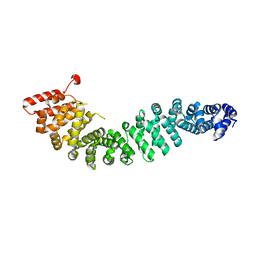 | | Crystal Structure of Beta-Catenin Armadillo Repeat Region and Its C-Terminal domain | | Descriptor: | Catenin beta-1 | | Authors: | Xing, Y, Takemaru, K, Liu, J, Zheng, J, Moon, R, Xu, W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-Length beta-Catenin
Structure, 16, 2008
|
|
2Z6G
 
 | | Crystal Structure of a Full-Length Zebrafish Beta-Catenin | | Descriptor: | B-catenin | | Authors: | Xing, Y, Takemaru, K, Liu, J, Zheng, J, Moon, R, Xu, W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of a Full-Length beta-Catenin
Structure, 16, 2008
|
|
1ZXC
 
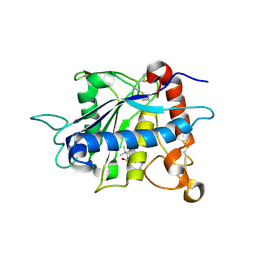 | | Crystal structure of catalytic domain of TNF-alpha converting enzyme (TACE) with inhibitor | | Descriptor: | (3S)-4-{[4-(BUT-2-YNYLOXY)PHENYL]SULFONYL}-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Zhang, Y, Jin, G, Cowling, R, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylenic TACE inhibitors. Part 2: SAR of six-membered cyclic sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A8H
 
 | | Crystal structure of catalytic domain of TACE with Thiomorpholine Sulfonamide Hydroxamate inhibitor | | Descriptor: | 4-({4-[(4-AMINOBUT-2-YNYL)OXY]PHENYL}SULFONYL)-N-HYDROXY-2,2-DIMETHYLTHIOMORPHOLINE-3-CARBOXAMIDE, ADAM 17, ZINC ION | | Authors: | Levin, J.I, Chen, J.M, Laakso, L.M, Du, M, Schmid, J, Xu, W, Cummons, T, Xu, J, Jin, G, Barone, D, Skotnicki, J.S. | | Deposit date: | 2005-07-08 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acetylenic TACE inhibitors. Part 3: Thiomorpholine sulfonamide hydroxamates.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
484D
 
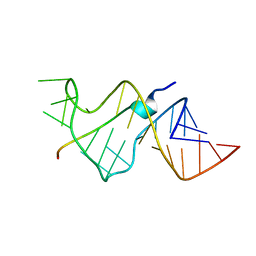 | | SOLUTION STRUCTURE OF HIV-1 REV PEPTIDE-RNA APTAMER COMPLEX | | Descriptor: | BASIC REV PEPTIDE, RNA APTAMER | | Authors: | Ye, X, Gorin, A.A, Frederick, R, Hu, W, Majumdar, A, Xu, W, Mclendon, G, Ellington, A, Patel, D.J. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA architecture dictates the conformations of a bound peptide.
Chem.Biol., 6, 1999
|
|
1Z87
 
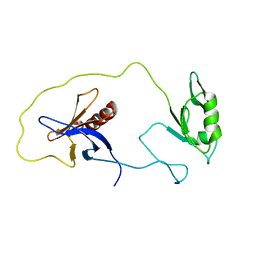 | | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1Z86
 
 | | Solution structure of the PDZ domain of alpha-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Xu, W, Wen, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-03-30 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
2ADZ
 
 | | solution structure of the joined PH domain of alpha1-syntrophin | | Descriptor: | Alpha-1-syntrophin | | Authors: | Yan, J, Wen, W, Xu, W, Long, J.F, Adams, M.E, Froehner, S.C, Zhang, M. | | Deposit date: | 2005-07-21 | | Release date: | 2006-01-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure of the split PH domain and distinct lipid-binding properties of the PH-PDZ supramodule of alpha-syntrophin
Embo J., 24, 2005
|
|
1G3J
 
 | | CRYSTAL STRUCTURE OF THE XTCF3-CBD/BETA-CATENIN ARMADILLO REPEAT COMPLEX | | Descriptor: | BETA-CATENIN ARMADILLO REPEAT REGION, TCF3-CBD (CATENIN BINDING DOMAIN) | | Authors: | Graham, T.A, Weaver, C, Mao, F, Kimelman, D, Xu, W. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a beta-catenin/Tcf complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
5CL1
 
 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
1SJY
 
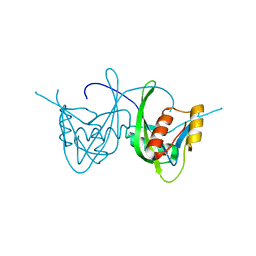 | | Crystal Structure of NUDIX HYDROLASE DR1025 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
5EJC
 
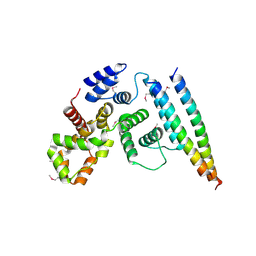 | | Crystal structural of the TSC1-TBC1D7 complex | | Descriptor: | Hamartin, TBC1 domain family member 7 | | Authors: | Wang, Z, Qin, J, Gong, W, Xu, W. | | Deposit date: | 2015-11-01 | | Release date: | 2016-03-02 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis of the Interaction between Tuberous Sclerosis Complex 1 (TSC1) and Tre2-Bub2-Cdc16 Domain Family Member 7 (TBC1D7).
J.Biol.Chem., 291, 2016
|
|
1SOI
 
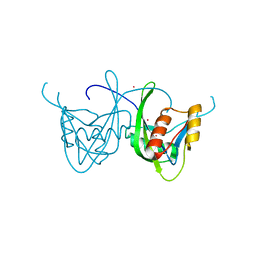 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEX WITH SM+3 | | Descriptor: | MutT/nudix family protein, SAMARIUM (III) ION | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
6UK5
 
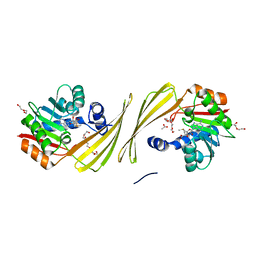 | | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis | | Descriptor: | ACETATE ION, CalS10, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alvarado, S.K, Miller, M.D, Xu, W, Wang, Z, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis
To Be Published
|
|
6UBL
 
 | | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina | | Descriptor: | DynF, PALMITIC ACID | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Bhardwaj, M, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2022-01-12 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
1SZ3
 
 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEXED WITH GNP AND MG+2 | | Descriptor: | MAGNESIUM ION, MutT/nudix family protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-04-02 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SU2
 
 | | CRYSTAL STRUCTURE OF THE NUDIX HYDROLASE DR1025 IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
6DA9
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6VJV
 
 | | Crystal structure of the Prochlorococcus phage (myovirus P-SSM2) ferredoxin at 1.6 Angstroms | | Descriptor: | ACETATE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Olmos Jr, J.L, Campbell, I.J, Miller, M.D, Xu, W, Kahanda, D, Atkinson, J.T, Sparks, N, Bennett, G.N, Silberg, J.J, Phillips Jr, G.N. | | Deposit date: | 2020-01-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Prochlorococcusphage ferredoxin: structural characterization and electron transfer to cyanobacterial sulfite reductases.
J.Biol.Chem., 295, 2020
|
|
6EF4
 
 | | Crystal structure of mouse PP2A Aalpha P179R mutant | | Descriptor: | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | | Authors: | Wang, Z, Shen, G, Xu, W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Highly Recurrent PP2A A alpha-Subunit Mutation P179R Alters Protein Structure and Impairs PP2A Enzyme Function to Promote Endometrial Tumorigenesis.
Cancer Res., 79, 2019
|
|
