7C38
 
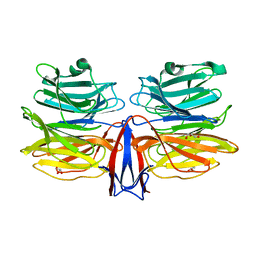 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with L-fucose | | Descriptor: | AofleA, GLYCEROL, alpha-L-fucopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C3E
 
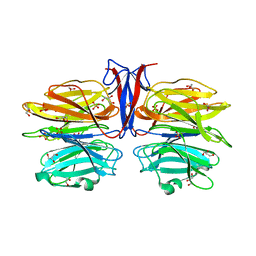 | | Crystal structure of R97A/R150A/R203A mutant of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, GLYCEROL, SULFATE ION | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C3C
 
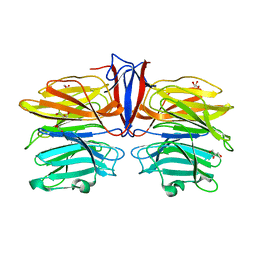 | | Crystal structure of AofleA from Arthrobotrys oligospora in complex with D-manose | | Descriptor: | AofleA, GLYCEROL, alpha-D-mannopyranose, ... | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-12 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
7C37
 
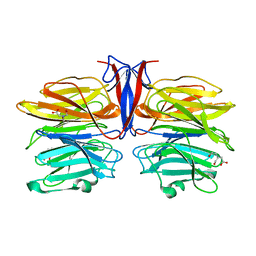 | | Crystal structure of AofleA from Arthrobotrys oligospora | | Descriptor: | AofleA, BICINE | | Authors: | Liu, M, Cheng, X, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the fungi-nematodes interaction mediated by fucose-specific lectin AofleA from Arthrobotrys oligospora.
Int.J.Biol.Macromol., 164, 2020
|
|
5JDS
 
 | |
5JDR
 
 | | Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zhou, A, Wei, H. | | Deposit date: | 2016-04-17 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade.
Cell Discov, 3, 2017
|
|
7EPC
 
 | | Cryo-EM structure of inactive mGlu7 homodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPA
 
 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPB
 
 | | Cryo-EM structure of LY354740-bound mGlu2 homodimer | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, Anti-RON nanobody, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPF
 
 | | Crystal structure of mGlu2 bound to NAM597 | | Descriptor: | (8~{R})-4-[2,4-bis(fluoranyl)phenyl]-8-methyl-7-[(2-methylpyrazol-3-yl)methyl]-6,8-dihydro-5~{H}-1,7-naphthyridine-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPD
 
 | | Cryo-EM structure of inactive mGlu2-7 heterodimer | | Descriptor: | Isoform 3 of Metabotropic glutamate receptor 7, Metabotropic glutamate receptor 2,Peptidylprolyl isomerase | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7EPE
 
 | | Crystal structure of mGlu2 bound to NAM563 | | Descriptor: | 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, FLAVIN MONONUCLEOTIDE, Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Lin, S, Han, S, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7JIJ
 
 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Powell, K, Xu, T, Brunzelle, J.S, Xu, H.X, Melcher, K. | | Deposit date: | 2020-07-23 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
7V3Z
 
 | | Structure of cannabinoid receptor type 1(CB1) | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, CHOLESTEROL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liu, Z.J, Shen, L, Hua, T, Yao, D.Q, Wu, L.J. | | Deposit date: | 2021-08-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | A Genetically Encoded F-19 NMR Probe Reveals the Allosteric Modulation Mechanism of Cannabinoid Receptor 1.
J.Am.Chem.Soc., 143, 2021
|
|
6KMH
 
 | | The crystal structure of CASK/Mint1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Li, W, Feng, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
6N4E
 
 | | hPGDS complexed with a quinoline-3-carboxamide | | Descriptor: | 7-(difluoromethoxy)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]quinoline-3-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase | | Authors: | Shewchuk, L.M, Ward, P. | | Deposit date: | 2018-11-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors.
Bioorg. Med. Chem., 27, 2019
|
|
6N69
 
 | | rat hPGDS complexed with a quinoline | | Descriptor: | GLUTATHIONE, Hematopoietic prostaglandin D synthase, quinoline-3-carbonitrile | | Authors: | Shewchuk, L.M, Cleasby, A. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors.
Bioorg. Med. Chem., 27, 2019
|
|
8H6P
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8H6T
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective small molecule inhibitor | | Descriptor: | (1R,3S)-3-{3-[(pyridin-2-yl)amino]-1H-pyrazol-5-yl}cyclopentyl propan-2-ylcarbamate, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8HEF
 
 | | The Crystal structure of deuterated S-217622 (Ensitrelvir) bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Synthesis of deuterated S-217622 (Ensitrelvir) with antiviral activity against coronaviruses including SARS-CoV-2.
Antiviral Res., 213, 2023
|
|
5XXY
 
 | |
7E2U
 
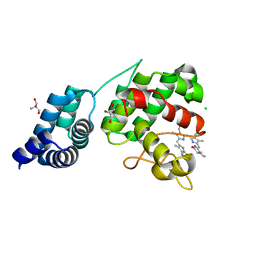 | | Synechocystis GUN4 in complex with phytochrome | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2R
 
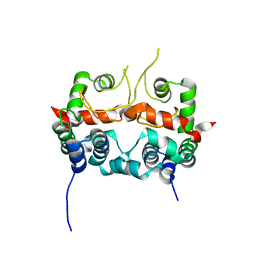 | | The ligand-free structure of Arabidopsis thaliana GUN4 | | Descriptor: | Tetrapyrrole-binding protein, chloroplastic | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2T
 
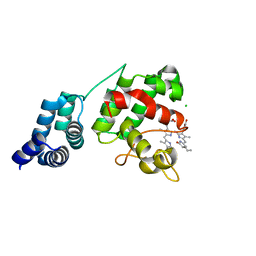 | | Synechocystis GUN4 in complex with phycocyanobilin | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2S
 
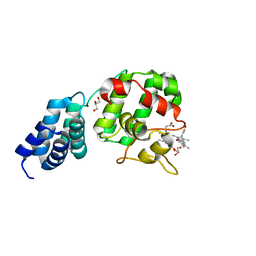 | | Synechocystis GUN4 in complex with biliverdin IXa | | Descriptor: | BILIVERDINE IX ALPHA, GLYCEROL, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
