8DH3
 
 | |
8DH5
 
 | |
8DH0
 
 | |
8EWG
 
 | | Cryo-EM structure of a riboendonclease | | 分子名称: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | 著者 | Li, Z, Wang, F. | | 登録日 | 2022-10-23 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
4M6T
 
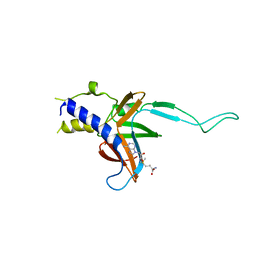 | | Structure of human Paf1 and Leo1 complex | | 分子名称: | RNA polymerase II-associated factor 1 homolog, Linker, RNA polymerase-associated protein LEO1, ... | | 著者 | Shen, Y, Qin, X. | | 登録日 | 2013-08-11 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | Structural insights into Paf1 complex assembly and histone binding
Nucleic Acids Res., 41, 2013
|
|
7KIB
 
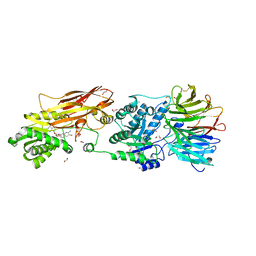 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 4 | | 分子名称: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Palte, R.L, Hayes, R.P. | | 登録日 | 2020-10-23 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7KID
 
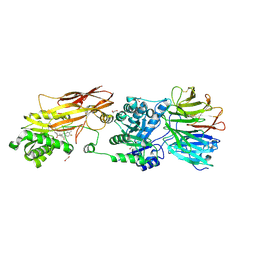 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 72 | | 分子名称: | (1S,2R,3aR,4S,6aR)-4-[(2-amino-3,5-difluoroquinolin-7-yl)methyl]-2-(4-amino-5-fluoro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydropentalene-1,6a(1H)-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | 著者 | Palte, R.L. | | 登録日 | 2020-10-23 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7KIC
 
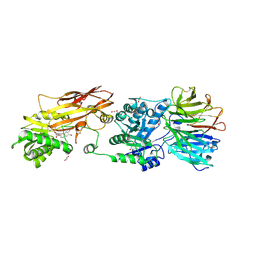 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 34 | | 分子名称: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | 著者 | Palte, R.L. | | 登録日 | 2020-10-23 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
3QJ8
 
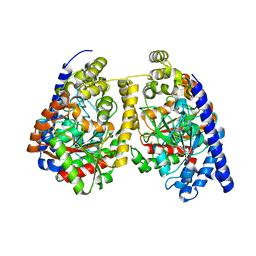 | |
7KQR
 
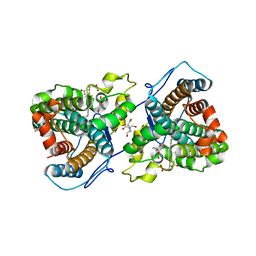 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | 著者 | Wang, Y, Shin, I, Liu, A. | | 登録日 | 2020-11-17 | | 公開日 | 2021-03-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | 著者 | Wang, Y, Liu, A. | | 登録日 | 2020-11-17 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.677 Å) | | 主引用文献 | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | 著者 | Wang, Y, Davis, I, Liu, A. | | 登録日 | 2020-11-17 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.579 Å) | | 主引用文献 | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
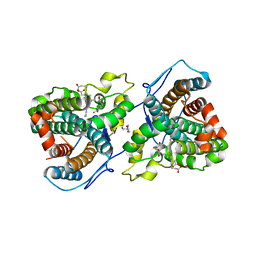 | |
3QKV
 
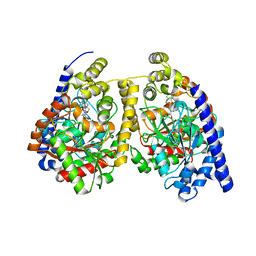 | | Crystal structure of fatty acid amide hydrolase with small molecule compound | | 分子名称: | (6-bromo-1'H,4H-spiro[1,3-benzodioxine-2,4'-piperidin]-1'-yl)methanol, Fatty-acid amide hydrolase 1 | | 著者 | Min, X, Walker, N.P.C, Wang, Z. | | 登録日 | 2011-02-01 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QJ9
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 1-{(3S)-1-[4-(1-benzofuran-2-yl)pyrimidin-2-yl]piperidin-3-yl}-3-ethyl-1,3-dihydro-2H-benzimidazol-2-one, Fatty-acid amide hydrolase 1, ... | | 著者 | Min, X, Walker, N.P.C, Wang, Z. | | 登録日 | 2011-01-28 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6JQ1
 
 | |
2X6L
 
 | |
2X69
 
 | |
2X6G
 
 | |
6CBV
 
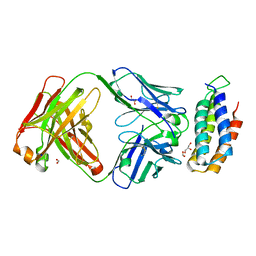 | | Crystal structure of BRIL bound to an affinity matured synthetic antibody. | | 分子名称: | BRIL, FORMIC ACID, GLYCEROL, ... | | 著者 | Mukherjee, S, Skrobek, B, Kossiakoff, A.A. | | 登録日 | 2018-02-05 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.872 Å) | | 主引用文献 | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
7QV2
 
 | |
7QV3
 
 | |
7QV1
 
 | |
7S4E
 
 | |
6L0O
 
 | | WH domain of human MCM8 | | 分子名称: | DNA helicase MCM8, SULFATE ION | | 著者 | Liu, Y, Li, J, Liu, L, Zeng, H. | | 登録日 | 2019-09-26 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Crystal structure of the winged-helix domain of MCM8.
Biochem.Biophys.Res.Commun., 526, 2020
|
|
