7EKX
 
 | |
6IVA
 
 | |
6IWW
 
 | | Cryo-EM structure of the S. typhimurium oxaloacetate decarboxylase beta-gamma sub-complex | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Oxaloacetate decarboxylase beta chain, Probable oxaloacetate decarboxylase gamma chain | | Authors: | Xu, X, Shi, H, Zhang, X, Xiang, S. | | Deposit date: | 2018-12-08 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into sodium transport by the oxaloacetate decarboxylase sodium pump.
Elife, 9, 2020
|
|
5D0F
 
 | |
5D06
 
 | |
2NQN
 
 | | MoeA T100W | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQR
 
 | | MoeA D142N | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQV
 
 | | MoeA D228A | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nciolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQK
 
 | | MoeA D59N mutant | | Descriptor: | Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQM
 
 | | MoeA T100A mutant | | Descriptor: | Molybdopterin biosynthesis protein moeA | | Authors: | Nicoas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQS
 
 | | MoeA E188A | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQU
 
 | | MoeA E188Q | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NQQ
 
 | | MoeA R137Q | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NRS
 
 | | MoeA S371W | | Descriptor: | Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-11-02 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NRP
 
 | | MoeA R350A | | Descriptor: | GLYCEROL, Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-11-02 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
2NRO
 
 | | MoeA K279Q | | Descriptor: | Molybdopterin biosynthesis protein moeA | | Authors: | Nicolas, J, Xiang, S, Schindelin, H, Rajagopalan, K.V. | | Deposit date: | 2006-11-02 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutational Analysis of Escherichia coli MoeA: Two Functional Activities Map to the Active Site Cleft.
Biochemistry, 46, 2007
|
|
5ZT0
 
 | |
5ZQV
 
 | |
2QL6
 
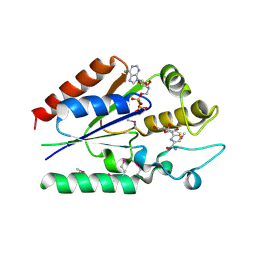 | | human nicotinamide riboside kinase (NRK1) | | Descriptor: | (1R)-1-[4-(AMINOCARBONYL)-1,3-THIAZOL-2-YL]-1,4-ANHYDRO-D-RIBITOL, ADENOSINE-5'-DIPHOSPHATE, nicotinamide riboside kinase 1 | | Authors: | Khan, J.A, Xiang, S, Tong, L. | | Deposit date: | 2007-07-12 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human nicotinamide riboside kinase
Structure, 15, 2007
|
|
2QG6
 
 | |
6E40
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complexed with ferric heme and Epacadostat | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E42
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and 4-Chlorophenyl imidazole | | Descriptor: | 4-(3-chlorophenyl)-1H-imidazole, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E41
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and an Epacadostat analog | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]sulfanyl}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E43
 
 | | Crystal structure of human indoleamine 2,3-dioxygenase 1 (IDO1) in complex with a BMS-978587 analog | | Descriptor: | (1R,2S)-2-(4-[cyclohexyl(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, BENZOIC ACID, Indoleamine 2,3-dioxygenase 1 | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E45
 
 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferrous state | | Descriptor: | GLYCEROL, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
