4MWY
 
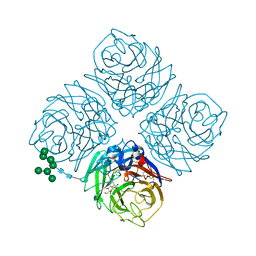 | | Shanghai N9-laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4NHG
 
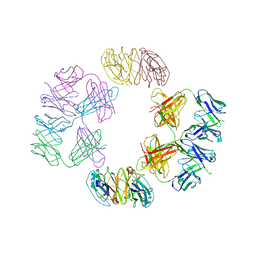 | | Crystal Structure of 2G12 IgG Dimer | | Descriptor: | 2G12 IgG dimer heavy chain, 2G12 IgG dimer light chain, Hepatitis B virus receptor binding protein | | Authors: | Wu, Y, West Jr, A.P, Kim, H.J, Thornton, M.E, Ward, A.B, Bjorkman, P.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12.
Cell Rep, 5, 2013
|
|
4NHH
 
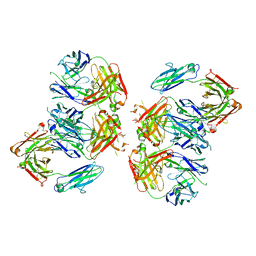 | | Structure of 2G12 IgG Dimer | | Descriptor: | 2G12 IgG dimer heavy chain, 2G12 IgG dimer light chain, Hepatitis B virus receptor binding protein | | Authors: | Wu, Y, West Jr, A.P, Kim, H.J, Thornton, M.E, Ward, A.B, Bjorkman, P.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12.
Cell Rep, 5, 2013
|
|
5Y10
 
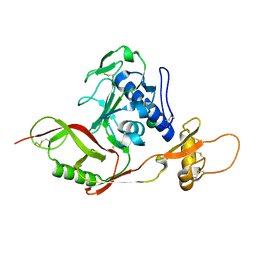 | | SFTSV Gn head domain | | Descriptor: | Membrane glycoprotein polyprotein, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wu, Y, Gao, F, Qi, J.X, Chai, Y, Gao, G.F. | | Deposit date: | 2017-07-19 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y0Y
 
 | | RVFV GN-AU | | Descriptor: | GOLD ION, NSmGnGc | | Authors: | Wu, Y, Gao, F, Qi, J.X, Chai, Y, Gao, G.F. | | Deposit date: | 2017-07-19 | | Release date: | 2017-09-13 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y11
 
 | | SFTSV GN with neutralizing antibody MAb4-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAb 4-5 heavy chain, MAb 4-5 light chain, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Chai, Y, Gao, G.F. | | Deposit date: | 2017-07-19 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y0W
 
 | | The structure of RVFV Gn head domain | | Descriptor: | NSmGnGc | | Authors: | Wu, Y, Gao, F, Qi, J.X, Chai, Y, Gao, G.F. | | Deposit date: | 2017-07-19 | | Release date: | 2017-09-13 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of phlebovirus glycoprotein Gn and identification of a neutralizing antibody epitope
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V8W
 
 | | Crystal structure of human Integrator IntS9-IntS11 CTD complex | | Descriptor: | Integrator complex subunit 11, Integrator complex subunit 9 | | Authors: | Wu, Y, Tong, L. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6LUP
 
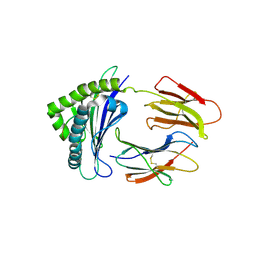 | | Crystal structure of shark MHC CLASS I for 2.3 angstrom | | Descriptor: | Beta-2-microglobulin, MHC class I protein, PHE-ALA-ASN-PHE-PHE-ILE-ARG-GLY-LEU | | Authors: | Wu, Y, Xia, C. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
7V51
 
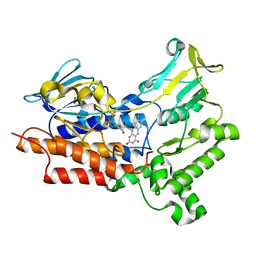 | | BVMO_negative mutant D432V | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H.-L. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7F0I
 
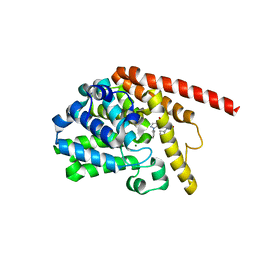 | | phosphodiesterase-9A in complex with inhibitor 4b | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(6-fluoranyl-2-azaspiro[3.3]heptan-2-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, Isoform PDE9A2 of High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.70000887 Å) | | Cite: | Discovery of Potent Phosphodiesterase-9 Inhibitors for the Treatment of Hepatic Fibrosis
J.Med.Chem., 64, 2021
|
|
7V4X
 
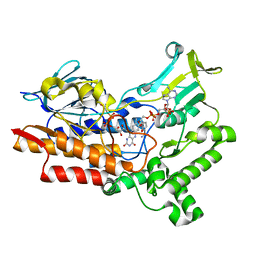 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-15 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V50
 
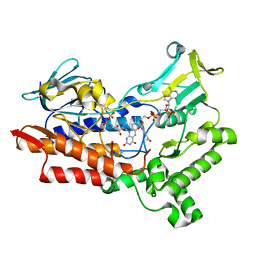 | | Structure of cyclohexanone monooxygenase mutant from Acinetobacter calcoaceticus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative flavin-binding monooxygenase | | Authors: | Wu, Y, Yu, H. | | Deposit date: | 2021-08-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7VHR
 
 | | Apostichopus japonicus ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Wu, Y, Su, X.R, Ming, T.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystallographic characterization of a marine invertebrate ferritin from the sea cucumber Apostichopus japonicus.
Febs Open Bio, 12, 2022
|
|
7Y74
 
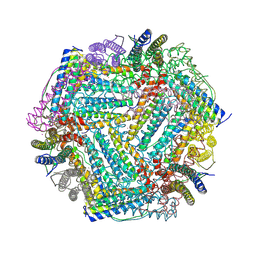 | |
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8BOB
 
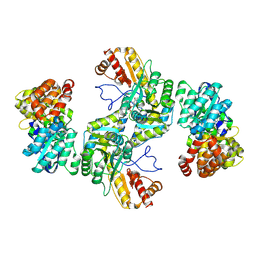 | | Structural basis for negative regulation of the maltose system | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HTH-type transcriptional regulator MalT, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Chai, J, Wu, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for negative regulation of the Escherichia coli maltose system.
Nat Commun, 14, 2023
|
|
5JQ9
 
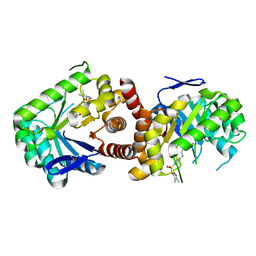 | | Yersinia pestis DHPS with pterine-sulfa conjugate Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-N-(3,4-dimethyl-1,2-oxazol-5-yl)benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Wu, Y, White, S.W. | | Deposit date: | 2016-05-04 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Pterin-sulfa conjugates as dihydropteroate synthase inhibitors and antibacterial agents.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2FPL
 
 | | RadA recombinase in complex with AMP-PNP and low concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2FPK
 
 | | RadA recombinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2FPM
 
 | | RadA recombinase in complex with AMP-PNP and high concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Methanoccocus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
2GDJ
 
 | | Delta-62 RADA recombinase in complex with AMP-PNP and magnesium | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wu, Y, Qian, X, He, Y, Luo, Y. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Rad51/RadA N-Terminal Domain Activates Nucleoprotein Filament ATPase Activity.
Structure, 14, 2006
|
|
5LE2
 
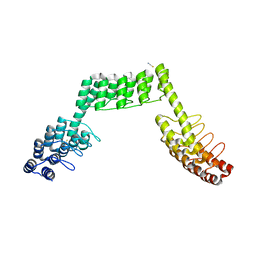 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_15_D12_15_D12 | | Descriptor: | ACETATE ION, DDD_D12_15_D12_15_D12, THIOCYANATE ION | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEC
 
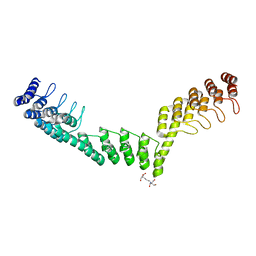 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DDD_D12_12_D12_12_D12 | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEL
 
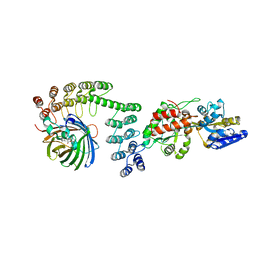 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_10_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | Descriptor: | DD_Off7_10_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | Authors: | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | Deposit date: | 2016-06-30 | | Release date: | 2017-11-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
