2JOY
 
 | | NMR Structure of 50S Ribosomal Protein L14e from Sulfolobus Solfataricus: Northeast Structural Genomics Consortium Target SSR105 | | Descriptor: | 50S ribosomal protein L14e | | Authors: | Singarapu, K.K, Wu, Y, Yee, A, Semesi, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of 50S Ribosomal Protein L14e; Northeast Structural Genomics Consortium Target SSR105
To be Published
|
|
2KI8
 
 | | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium target AtT7 | | Descriptor: | Tungsten formylmethanofuran dehydrogenase, subunit D (FwdD-2) | | Authors: | Eletsky, A, Wu, Y, Yee, A, Fares, C, Lee, H.W, Arrowsmith, C.H, Prestegard, J.H, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of tungsten formylmethanofuran dehydrogenase subunit D from Archaeoglobus fulgidus
To be Published
|
|
2KC7
 
 | | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218 | | Descriptor: | bfr218_protein | | Authors: | Eletsky, A, Wu, Y, Sukumaran, D, Lee, H, Lee, D.Y, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-12-17 | | Release date: | 2009-01-13 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Bacteroides fragilis protein BF1650. Northeast Structural Genomics Consortium target BfR218
To be Published
|
|
2K4Y
 
 | | NMR Structure of FeoA-like protein from Clostridium acetobutylicum: Northeast Structural Genomics Consortium Target CaR178 | | Descriptor: | FeoA-like protein | | Authors: | Singarapu, K, Wu, Y, Hua, J, Sukumaran, D, Zhao, L, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of FeoA-like protein from Clostridium acetobutylicum: Northeast Structural Genomics Consortium Target CaR178
To be Published
|
|
2K5L
 
 | | Solution NMR Structure of Protein FeoA from Clostridium thermocellum, Northeast Structural Genomics Consortium Target CmR17 | | Descriptor: | FeoA | | Authors: | Zeri, A, Singarapu, K.K, Mills, J.L, Wu, Y, Garcia, E, Wang, H, Jiang, M, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-29 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Protein FeoA from
Clostridium thermocellum, Northeast Structural Genomics Consortium Target CmR17
To be Published
|
|
2KKV
 
 | | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H | | Descriptor: | Integrase | | Authors: | Mills, J.L, Wu, Y, Sukumaran, D.K, Belote, R.L, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G.V.T, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of an integrase domain from protein SPA4288 from Salmonella enterica, Northeast Structural Genomics Consortium Target SlR105H
To be Published
|
|
2KZ2
 
 | | Calmodulin, C-terminal domain, F92E mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2LFF
 
 | | Solution structure of Diiron protein in presence of 8 eq Zn2+, Northeast Structural Genomics consortium target OR21 | | Descriptor: | Diiron protein, ZINC ION | | Authors: | Pires, M, Wu, Y, Mills, J.L, Reig, A, Englander, W, Degrado, W, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Diiron protein in presence of 8 eq Zn2+
To be Published
|
|
2MLD
 
 | |
2M01
 
 | | Solution structure of Kunitz-type neurotoxin LmKKT-1a from scorpion venom | | Descriptor: | Protease inhibitor LmKTT-1a | | Authors: | Luo, F, Jiang, L, Liu, M, Chen, Z, Wu, Y. | | Deposit date: | 2012-10-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins.
Plos One, 8, 2013
|
|
2MLA
 
 | |
2M3S
 
 | | Calmodulin, i85l, f92e, h107i, l112r, a128t, m144r mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Moroz, Y.S, Wu, Y, Cheng, H, Roder, H, Korendovych, I.V. | | Deposit date: | 2013-01-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A single mutation in a regulatory protein produces evolvable allosterically regulated catalyst of nonnatural reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2MUZ
 
 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
6GZC
 
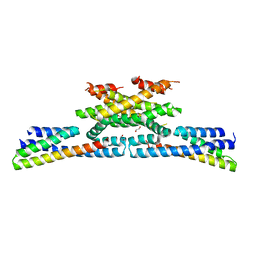 | | heterotetrameric katanin p60:p80 complex | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Katanin p60 ATPase-containing subunit A1, ... | | Authors: | Faltova, L, Jiang, K, Frey, D, Wu, Y, Capitani, G, Prota, A.E, Akhmanova, A, Steinmetz, M.O, Kammerer, R.A. | | Deposit date: | 2018-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Heterotetrameric Katanin p60:p80 Complex.
Structure, 27, 2019
|
|
6GWE
 
 | | Crystal structure of Thrombin bound to P2 macrocycle | | Descriptor: | (10S,14S,17R)-14-(3-carbamimidamidopropyl)-3-[[2-(hydroxymethyl)phenyl]methyl]-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(21),22,24-triene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cendron, L, Angelini, A, Kale, S.S, Bergeron-Brlek, M, Wu, Y, Heinis, C. | | Deposit date: | 2018-06-23 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thiol-to-amine cyclization reaction enables screening of large libraries of macrocyclic compounds and the generation of sub-kilodalton ligands.
Sci Adv, 5, 2019
|
|
6K87
 
 | |
6K82
 
 | | RGLG1 mutant-D338A E378A | | Descriptor: | E3 ubiquitin-protein ligase RGLG1, MAGNESIUM ION, SODIUM ION | | Authors: | Wang, Q, Wu, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | RGLG1 mutant-D338A E378A
To Be Published
|
|
6K89
 
 | | The closed state of RGLG1 VWA domain | | Descriptor: | E3 ubiquitin-protein ligase RGLG1, MAGNESIUM ION, SODIUM ION | | Authors: | Wang, Q, Wu, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | The closed state of RGLG1 VWA domain
To Be Published
|
|
6K88
 
 | | RGLG1 MIDAS binds calcium ion | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase RGLG1 | | Authors: | Wang, Q, Wu, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | RGLG1 MIDAS binds calcium ion
To Be Published
|
|
6K85
 
 | | The closed state of RGLG1 mutant-D338A | | Descriptor: | E3 ubiquitin-protein ligase RGLG1, MAGNESIUM ION | | Authors: | Wang, Q, Wu, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | The closed state of RGLG1 mutant-D338A
To Be Published
|
|
6K8A
 
 | |
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4HNM
 
 | | Crystal structure of human catenin-beta-like 1 56 kDa fragment | | Descriptor: | Beta-catenin-like protein 1 | | Authors: | Du, Z, Huang, X, Wang, G, Wu, Y. | | Deposit date: | 2012-10-19 | | Release date: | 2013-07-31 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.9001 Å) | | Cite: | The structure of full-length human CTNNBL1 reveals a distinct member of the armadillo-repeat protein family.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8H69
 
 | | Cryo-EM structure of influenza RNA polymerase | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*UP*AP*AP*AP*CP*UP*CP*CP*UP*GP*CP*UP*UP*UP*UP*GP*CP*U)-3'), ... | | Authors: | Li, H, Wu, Y, Liang, H, Liu, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4HM9
 
 | | Crystal structure of full-length human catenin-beta-like 1 | | Descriptor: | Beta-catenin-like protein 1 | | Authors: | Du, Z, Huang, X, Wang, G, Wu, Y. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | The structure of full-length human CTNNBL1 reveals a distinct member of the armadillo-repeat protein family.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
