8KA5
 
 | |
8KA4
 
 | |
3LON
 
 | | HCV NS3-4a protease domain with ketoamide inhibitor narlaprevir | | 分子名称: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, BETA-MERCAPTOETHANOL, Genome polyprotein, ... | | 著者 | Prongay, A.J. | | 登録日 | 2010-02-04 | | 公開日 | 2011-01-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Candidate selection and preclinical evaluation culminating in the discovery of Narlaprevir (SCH 900518): A potent, selective and orally efficacious second generation HCV NS3 serine protease inhibitor
To be Published
|
|
6OT1
 
 | |
6OSY
 
 | |
2NAE
 
 | |
6W03
 
 | |
1C9Q
 
 | |
3KLX
 
 | |
3KL1
 
 | |
4X32
 
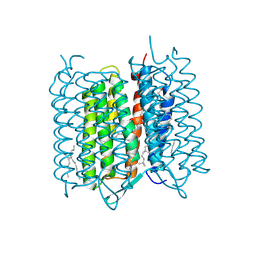 | | Bacteriorhodopsin ground state structure collected in cryo conditions from crystals obtained in LCP with PEG as a precipitant. | | 分子名称: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | 著者 | Nogly, P, Standfuss, J. | | 登録日 | 2014-11-27 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Lipidic cubic phase serial millisecond crystallography using synchrotron radiation.
Iucrj, 2, 2015
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | 分子名称: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | 分子名称: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2020-02-26 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.254 Å) | | 主引用文献 | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
8TMA
 
 | |
8TM1
 
 | |
6VZI
 
 | |
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | 分子名称: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | 著者 | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | 登録日 | 2010-08-23 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
6IE2
 
 | | Crystal structure of methyladenine demethylase | | 分子名称: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | 著者 | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | 登録日 | 2018-09-13 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
6IE3
 
 | | Crystal structure of methyladenine demethylase | | 分子名称: | ETHANOL, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | 著者 | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | 登録日 | 2018-09-13 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
7RGW
 
 | |
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | 著者 | Zhang, X, Zhang, Q, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | 著者 | Zhang, X, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DS8
 
 | | Complex structure of abscisic acid receptor PYL3-(+)-ABA-HAB1 in the presence of Mn2+ | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, GLYCEROL, ... | | 著者 | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | 登録日 | 2012-02-18 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8FLN
 
 | | Crystal structure of BTK C481S kinase domain in complex with pirtobrutinib | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | 著者 | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | 登録日 | 2022-12-21 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.334 Å) | | 主引用文献 | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
8FLL
 
 | | Crystal structure of BTK kinase domain in complex with pirtobrutinib | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | 著者 | Cedervall, E.P, Morales, T.H, Allerston, C.K. | | 登録日 | 2022-12-21 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-07-12 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | Preclinical characterization of pirtobrutinib, a highly selective, noncovalent (reversible) BTK inhibitor.
Blood, 142, 2023
|
|
