7FD3
 
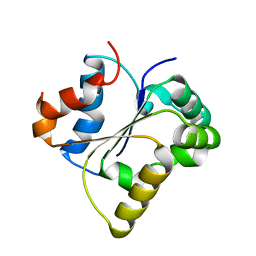 | | IL-1RAPL2 TIR domain | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Wang, X, Zhou, J. | | Deposit date: | 2021-07-15 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Iscience, 25, 2022
|
|
6M0J
 
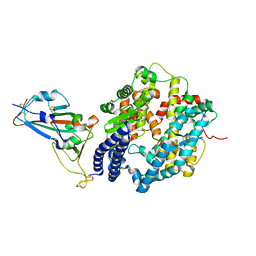 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
6K8K
 
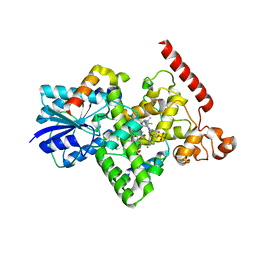 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7WTH
 
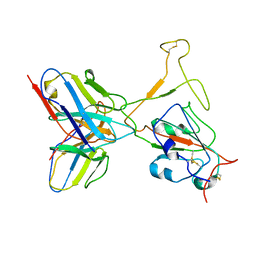 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv264 | | Descriptor: | Heavy chain of XGv264, Light chain of XGv264, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
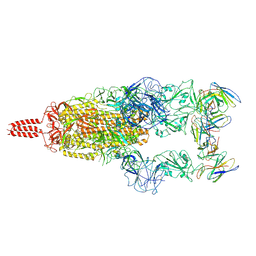 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTG
 
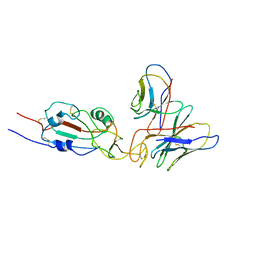 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv051 | | Descriptor: | Heavy chain of XGv051, Light chain of XGv051, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTJ
 
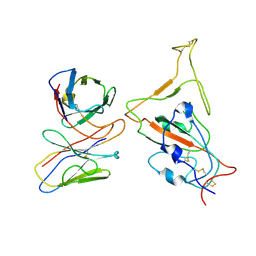 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv286 | | Descriptor: | Heavy chain of XGv286, Light chain of XGv286, Spike protein S1 | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTK
 
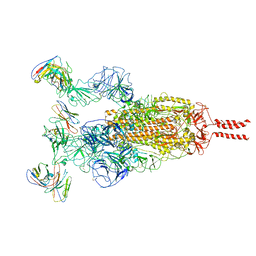 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTI
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv264 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv264, Light chain of XGv264, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
5YDG
 
 | | Crystal structure of the Arabidopsis thaliana chloroplast RNA editing factors 2(MORF2) | | Descriptor: | Multiple organellar RNA editing factor 2, chloroplastic | | Authors: | Wang, X, Yang, J.Y, Wang, Y.L, Gao, Y.S. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of the chloroplast RNA editing factor MORF2
Biochem. Biophys. Res. Commun., 495, 2018
|
|
7WT9
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab 9A8 | | Descriptor: | Heavy chain of Fab 9A8, Light chain of Fab 9A8, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT7
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT8
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7YKR
 
 | |
7YKS
 
 | |
7YF2
 
 | |
7YF3
 
 | |
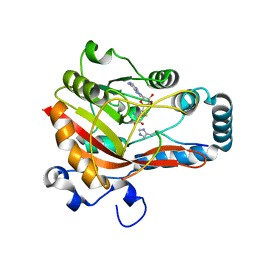
7Y9C
 
 | |
7YF4
 
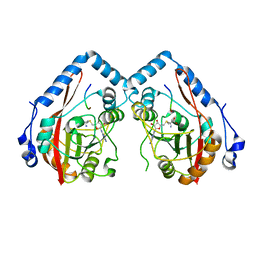 | |
5Y18
 
 | |
7CVN
 
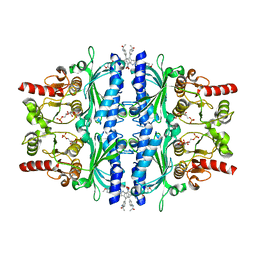 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-(3-acetamidophenyl)-N-(4-methoxyphenyl)sulfonyl-7-nitro-1H-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X, Zhou, J, Xu, B. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design,synthesis,biological evaluation and binding mode analysis of 7-nitro-indole-N-acylarylsulfonamide-based fructose-1,6-bisphosphatase inhibitors
Chinese journal of medicinal chemistry, 30, 2020
|
|
7CN8
 
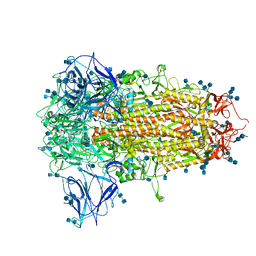 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN4
 
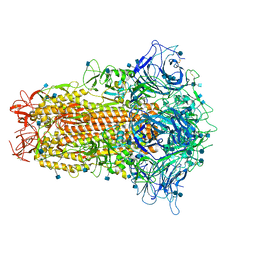 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7DHD
 
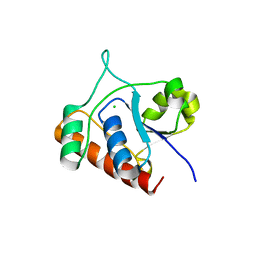 | | Vibrio vulnificus Wzb | | Descriptor: | CHLORIDE ION, Protein-tyrosine-phosphatase | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7DHF
 
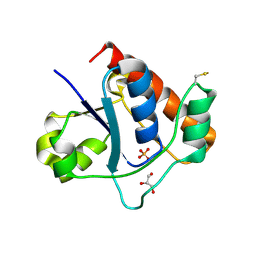 | | Vibrio vulnificus Wzb in complex with benzylphosphonate | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine-phosphatase | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
