2HAV
 
 | | Apo-Human Serum Transferrin (Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
2HAU
 
 | | Apo-Human Serum Transferrin (Non-Glycosylated) | | Descriptor: | CITRIC ACID, GLYCEROL, Serotransferrin | | Authors: | Wally, J, Everse, S.J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Iron-free Human Serum Transferrin Provides Insight into Inter-lobe Communication and Receptor Binding.
J.Biol.Chem., 281, 2006
|
|
5KK3
 
 | | Atomic Resolution Structure of Monomorphic AB42 Amyloid Fibrils | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Colvin, M.T, Silvers, R, Zhe Ni, Q, Can, T.V, Sergeyev, I, Rosay, M, Donovan, K.J, Michael, B, Wall, J, Linse, S, Griffin, R.G. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic Resolution Structure of Monomorphic A beta 42 Amyloid Fibrils.
J.Am.Chem.Soc., 138, 2016
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5W0M
 
 | | Structure of human TUT7 catalytic module (CM) in complex with U5 RNA | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W0B
 
 | | Structure of human TUT7 catalytic module (CM) | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-30 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W0O
 
 | |
5W0N
 
 | | Structure of human TUT7 catalytic module (CM) in complex with UMPNPP and U2 RNA | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5DXU
 
 | | p110delta/p85alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
5DXH
 
 | | p110alpha/p85alpha with compound 5 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, methyl {2-[4-(2-chlorophenyl)-4H-1,2,4-triazol-3-yl]-4,5-dihydrothieno[3,2-d][1]benzoxepin-8-yl}carbamate | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
5DXT
 
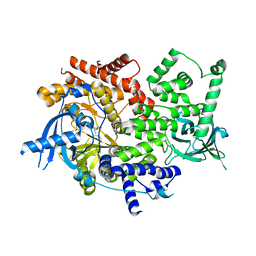 | | p110alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, 1,2-ETHANEDIOL, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
2MSR
 
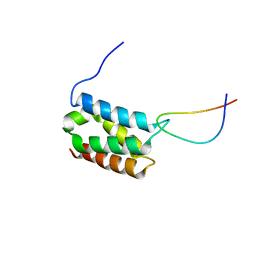 | | Solution structure of LEDGF/p75 IBD in complex with MLL1 peptide (140-160) | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein | | Authors: | Cermakova, K, Tesina, P, Demeulemeester, J, El Ashkar, S, Mereau, H, Schwaller, J, Rezacova, P, Veverka, V, De Rijck, J. | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Validation and Structural Characterization of the LEDGF/p75-MLL Interface as a New Target for the Treatment of MLL-Dependent Leukemia.
Cancer Res., 74, 2014
|
|
4PMW
 
 | |
6RZH
 
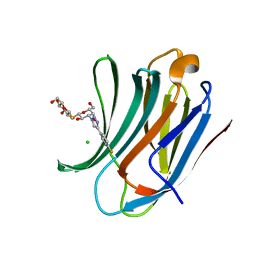 | | Galectin-3C in complex with para-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(4-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.947 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
6RZG
 
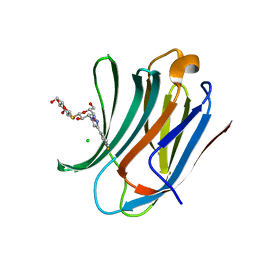 | | Galectin-3C in complex with meta-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(3-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.015 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
6RZF
 
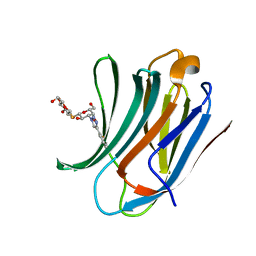 | | Galectin-3C in complex with ortho-fluoroaryltriazole galactopyranosyl 1-thio-D-glucopyranoside derivative | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-4-[4-(2-fluorophenyl)-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]sulfanyl-6-(hydroxymethyl)oxane-3,4,5-triol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2019-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.016 Å) | | Cite: | Entropy-Entropy Compensation between the Protein, Ligand, and Solvent Degrees of Freedom Fine-Tunes Affinity in Ligand Binding to Galectin-3C.
Jacs Au, 1, 2021
|
|
5N6Q
 
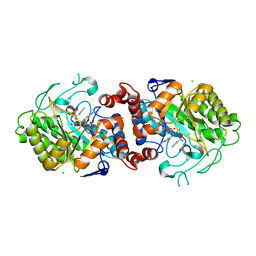 | | Xenobiotic reductase A (XenA) from Pseudomonas putida in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
5N6G
 
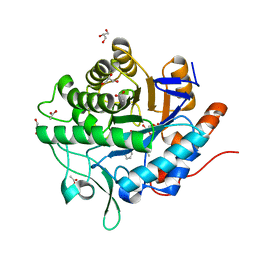 | | NerA from Agrobacterium radiobacter in complex with 2-phenylacrylic acid | | Descriptor: | 2-Phenylacrylic acid, ACETATE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural insights into the ene-reductase synthesis of profens.
Org. Biomol. Chem., 15, 2017
|
|
2NAO
 
 | | Atomic resolution structure of a disease-relevant Abeta(1-42) amyloid fibril | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Waelti, M.A, Ravotti, F, Arai, H, Glabe, C, Wall, J, Bockmann, A, Guntert, P, Meier, B.H, Riek, R. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Atomic-resolution structure of a disease-relevant A beta (1-42) amyloid fibril.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F1R
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (C10) | | Descriptor: | (3~{R})-8-cyclopropyl-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, Listeriolysin regulatory protein | | Authors: | Begum, A, Grundstrom, C, Good, J.A.D, Andersson, C, ALmqvist, F, Johansson, J, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Attenuating Listeria monocytogenes Virulence by Targeting the Regulatory Protein PrfA.
Cell Chem Biol, 23, 2016
|
|
6EXM
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK202) | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Grundstrom, C, Begum, A, Hall, M, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUT
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK67) | | Descriptor: | (3~{R})-8-cyclopropyl-7-[(4-methylnaphthalen-1-yl)methyl]-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUZ
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK37) | | Descriptor: | (3~{R})-8-methoxy-7-(naphthalen-1-ylmethyl)-5-oxidanylidene-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EXL
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (MK206) - folded HTH motif | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION, ... | | Authors: | Hall, M, Grundstrom, C, Begum, A, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-11-08 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
6EUU
 
 | | The Transcriptional Regulator PrfA from Listeria Monocytogenes in complex with a ring-fused 2-pyridone (KSK29) | | Descriptor: | (3~{R})-8-cyclopropyl-5-oxidanylidene-7-(quinolin-3-ylmethyl)-2,3-dihydro-[1,3]thiazolo[3,2-a]pyridine-3-carboxylic acid, ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, ... | | Authors: | Begum, A, Hall, M, Grundstrom, C, Kulen, M, Lindgren, M, Johansson, J, Almqvist, F, Sauer, U.H, Sauer-Eriksson, A.E. | | Deposit date: | 2017-10-31 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Targeting PrfA, the Master Virulence Regulator of Listeria monocytogenes.
J. Med. Chem., 61, 2018
|
|
