1MO3
 
 | |
1MO4
 
 | |
1MO5
 
 | |
2INS
 
 | | THE STRUCTURE OF DES-PHE B1 BOVINE INSULIN | | Descriptor: | DES-PHE B1 INSULIN (CHAIN A), DES-PHE B1 INSULIN (CHAIN B), ZINC ION | | Authors: | Smith, G.D, Duax, W.L, Dodson, E.J, Dodson, G.G, Degraaf, R.A.G, Reynolds, C.D. | | Deposit date: | 1982-05-10 | | Release date: | 1982-08-05 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of Des-Phe B1 Bovine Insulin
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1WBF
 
 | | WINGED BEAN LECTIN, SACCHARIDE FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Suguna, K. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of basic winged-bean lectin and a comparison with its saccharide-bound form.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3M3U
 
 | |
8I67
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 2,4-Thiazolidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 1,3-thiazolidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I61
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I69
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Fluoroorotic acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I6B
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, CHLORIDE ION, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I63
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I66
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid) and citric acid, Form I | | Descriptor: | 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, CITRIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I6D
 
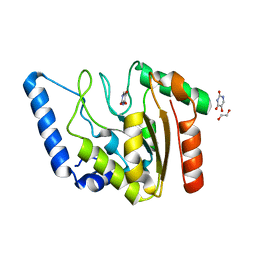 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form VI | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I62
 
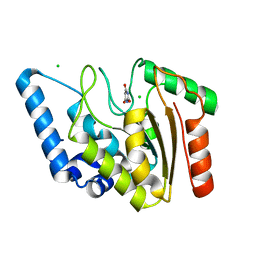 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, CHLORIDE ION, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I64
 
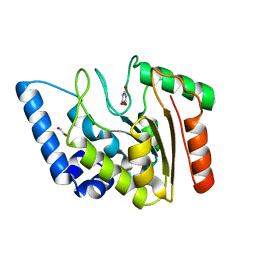 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid, Form II | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I68
 
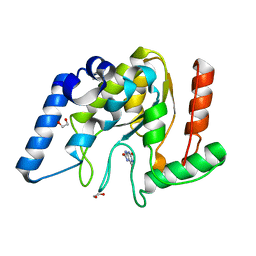 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Uric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, URIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I65
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with isoorotic acid (2,4-Dihydroxypyrimidine-5-carboxylic Acid), Form I | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carboxylic acid, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I6A
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Orotic acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, OROTIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
8I6C
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 6-Formyl-uracil, Form III | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment
Eur.J.Med.Chem., 258, 2023
|
|
