1I3J
 
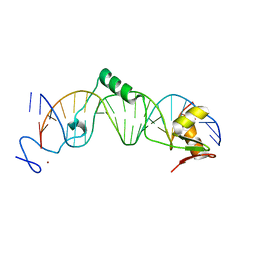 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF INTRON ENDONUCLEASE I-TEVI WITH ITS SUBSTRATE | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*AP*CP*GP*GP*TP*AP*GP*AP*CP*CP*CP*AP*AP*GP*A)-3', 5'-D(*TP*TP*CP*TP*TP*GP*GP*GP*TP*CP*TP*AP*CP*CP*GP*TP*TP*TP*AP*AP*T)-3', INTRON-ASSOCIATED ENDONUCLEASE 1, ... | | Authors: | Van Roey, P, Waddling, C.A, Fox, K.M, Belfort, M, Derbyshire, V. | | Deposit date: | 2001-02-15 | | Release date: | 2001-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate.
EMBO J., 20, 2001
|
|
1LN0
 
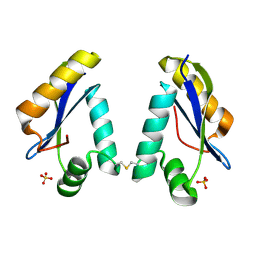 | | Structure of the Catalytic Domain of Homing Endonuclease I-TevI | | Descriptor: | SULFATE ION, intron-associated endonuclease 1 | | Authors: | Van Roey, P, Meehan, L, Kowalski, J.C, Belfort, M, Derbyshire, V. | | Deposit date: | 2002-05-02 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI.
Nat.Struct.Biol., 9, 2002
|
|
1MK0
 
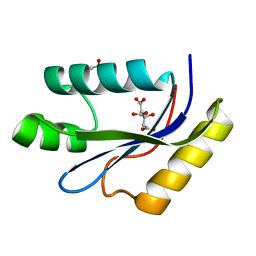 | | catalytic domain of intron endonuclease I-TevI, E75A mutant | | Descriptor: | BETA-MERCAPTOETHANOL, CITRIC ACID, Intron-associated endonuclease 1 | | Authors: | Van Roey, P, Meehan, L, Kowalski, J.C, Belfort, M, Derbyshire, V. | | Deposit date: | 2002-08-28 | | Release date: | 2002-10-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI.
Nat.Struct.Biol., 9, 2002
|
|
2EBN
 
 | |
2IN8
 
 | | crystal structure of Mtu recA intein, splicing domain | | Descriptor: | Endonuclease PI-MtuI, SULFATE ION | | Authors: | Van Roey, P. | | Deposit date: | 2006-10-06 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue.
J.Mol.Biol., 367, 2007
|
|
2IMZ
 
 | | Crystal structure of Mtu recA intein splicing domain | | Descriptor: | Endonuclease PI-MtuI, ZINC ION | | Authors: | Van Roey, P. | | Deposit date: | 2006-10-05 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue.
J.Mol.Biol., 367, 2007
|
|
2IN0
 
 | | crystal structure of Mtu recA intein splicing domain | | Descriptor: | Endonuclease PI-MtuI | | Authors: | Van Roey, P. | | Deposit date: | 2006-10-05 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue.
J.Mol.Biol., 367, 2007
|
|
2IN9
 
 | | crystal structure of Mtu recA intein, splicing domain | | Descriptor: | Endonuclease PI-MtuI, SULFATE ION | | Authors: | Van Roey, P. | | Deposit date: | 2006-10-06 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue.
J.Mol.Biol., 367, 2007
|
|
2MCM
 
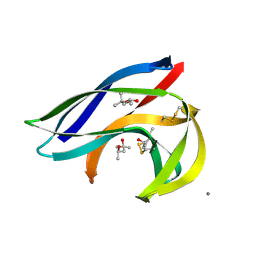 | | MACROMOMYCIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Van Roey, P. | | Deposit date: | 1991-05-08 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure analysis of auromomycin apoprotein (macromomycin) shows importance of protein side chains to chromophore binding selectivity.
Proc.Natl.Acad.Sci.Usa, 86, 1989
|
|
1PNF
 
 | | PNGASE F COMPLEX WITH DI-N-ACETYLCHITOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F, SULFATE ION | | Authors: | Van Roey, P, Kuhn, P. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site and oligosaccharide recognition residues of peptide-N4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase F.
J.Biol.Chem., 270, 1995
|
|
1PNG
 
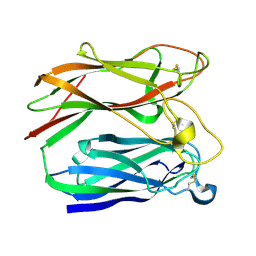 | |
1EDT
 
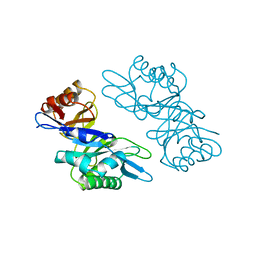 | |
1AYY
 
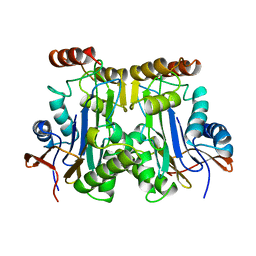 | | GLYCOSYLASPARAGINASE | | Descriptor: | GLYCOSYLASPARAGINASE | | Authors: | Van Roey, P, Xuan, J. | | Deposit date: | 1997-11-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of glycosylasparaginase from Flavobacterium meningosepticum.
Protein Sci., 7, 1998
|
|
1B24
 
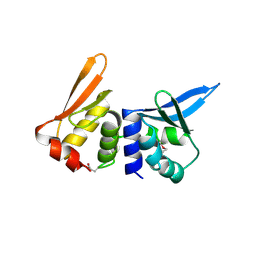 | | I-DMOI, INTRON-ENCODED ENDONUCLEASE | | Descriptor: | Homing endonuclease I-DmoI | | Authors: | Van Roey, P, Silva, G.H. | | Deposit date: | 1998-12-03 | | Release date: | 1999-03-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI.
J.Mol.Biol., 286, 1999
|
|
3NZM
 
 | |
3IFJ
 
 | |
3IGD
 
 | |
4V48
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4V47
 
 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the EF-G.GTP state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
1VQ2
 
 | | CRYSTAL STRUCTURE OF T4-BACTERIOPHAGE DEOXYCYTIDYLATE DEAMINASE, MUTANT R115E | | Descriptor: | 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, DEOXYCYTIDYLATE DEAMINASE, ZINC ION | | Authors: | Almog, R, Maley, F, Maley, G.F, Maccoll, R, Van Roey, P. | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of the R115E Mutant of T4-Bacteriophage 2'-Deoxycytidylate Deaminase
Biochemistry, 43, 2004
|
|
1HZW
 
 | | CRYSTAL STRUCTURE OF HUMAN THYMIDYLATE SYNTHASE | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Almog, R, Waddling, C.A, Maley, F, Maley, G.F, Van Roey, P. | | Deposit date: | 2001-01-26 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a deletion mutant of human thymidylate synthase Delta (7-29) and its ternary complex with Tomudex and dUMP.
Protein Sci., 10, 2001
|
|
1I00
 
 | | CRYSTAL STRUCTURE OF HUMAN THYMIDYLATE SYNTHASE, TERNARY COMPLEX WITH DUMP AND TOMUDEX | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE, TOMUDEX | | Authors: | Almog, R.A, Waddling, C.A, Maley, F, Maley, G.F, Van Roey, P. | | Deposit date: | 2001-01-27 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a deletion mutant of human thymidylate synthase Delta (7-29) and its ternary complex with Tomudex and dUMP.
Protein Sci., 10, 2001
|
|
1T2T
 
 | | Crystal structure of the DNA-binding domain of intron endonuclease I-TevI with operator site | | Descriptor: | 5'-D(*AP*AP*TP*TP*AP*AP*AP*GP*GP*GP*CP*AP*GP*TP*CP*CP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*TP*GP*TP*AP*GP*GP*AP*CP*TP*GP*CP*CP*CP*TP*TP*TP*AP*AP*T)-3', Intron-associated endonuclease 1, ... | | Authors: | Edgell, D.R, Derbyshire, V, Van Roey, P, LaBonne, S, Stanger, M.J, Li, Z, Boyd, T.M, Shub, D.A, Belfort, M. | | Deposit date: | 2004-04-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1L9X
 
 | | Structure of gamma-Glutamyl Hydrolase | | Descriptor: | BETA-MERCAPTOETHANOL, gamma-glutamyl hydrolase | | Authors: | Li, H, Ryan, T.J, Chave, K.J, Van Roey, P. | | Deposit date: | 2002-03-26 | | Release date: | 2002-04-10 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure of human gamma -glutamyl hydrolase. A class I glatamine amidotransferase adapted for a complex substate.
J.Biol.Chem., 277, 2002
|
|
1EOK
 
 | | CRYSTAL STRUCTURE OF ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3 | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE F3, SULFATE ION | | Authors: | Waddling, C.A, Plummer Jr, T.H, Tarentino, A.L, Van Roey, P. | | Deposit date: | 2000-03-23 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3).
Biochemistry, 39, 2000
|
|
