7EQE
 
 | | Crystal Structure of a transcription factor | | Descriptor: | TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
7EQF
 
 | | Crystal Structure of a Transcription Factor in complex with Ligand | | Descriptor: | (6~{R})-3-methyl-8-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{R},5~{R},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-[(2~{S},4~{R},5~{S},6~{R})-6-methyl-5-[(2~{S},4~{S},5~{S},6~{R})-6-methyl-4-[(2~{S},5~{S},6~{S})-6-methyl-5-oxidanyl-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-oxan-2-yl]oxy-5-oxidanyl-oxan-2-yl]oxy-4-oxidanyl-oxan-2-yl]oxy-1,6,11-tris(oxidanyl)-5,6-dihydrobenzo[a]anthracene-7,12-dione, TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
9LVT
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 4 | | Descriptor: | 1-(2-azanylideneethyl)-6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-5-[[3,4,5-tris(fluoranyl)phenyl]methyl]pyrimidine-2,4-dione, 3C-like proteinase | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
9LVV
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 17 (S-892216) | | Descriptor: | 1-(2-azanylideneethyl)-6-[6,6-bis(fluoranyl)-2-azaspiro[3.3]heptan-2-yl]-5-(3-chloranyl-4-fluoranyl-phenyl)-3-(5-chloranylpyridin-3-yl)pyrimidine-2,4-dione, 3C-like proteinase | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
9LVR
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, 6-(1,3-dihydroisoindol-2-yl)-3-(5-methylpyridin-3-yl)-1-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Unoh, Y, Hirai, K, Uehara, S, Kawashima, S, Nobori, H, Sato, J, Shibayama, H, Hori, A, Nakahara, K, Kurahashi, K, Takamatsu, M, Yamamoto, S, Zhang, O, Tanimura, M, Dodo, R, Maruyama, Y, Sawa, H, Watari, R, Miyano, T, Kato, T, Sato, T, Tachibana, Y. | | Deposit date: | 2025-02-12 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of The Clinical Candidate S-892216: A Second-Generation of SARS-CoV-2 3CL Protease Inhibitor for Treating COVID-19
To Be Published
|
|
7WMC
 
 | | Crystal structure of macrocyclic peptide 1 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, Peptide1 | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
7WMT
 
 | | Crystal structure of small molecule 13 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, [(2~{R},4~{S})-4-[2-(aminomethyl)imidazol-1-yl]-2-[1-[(4-chlorophenyl)methyl]-5-methyl-indol-2-yl]pyrrolidin-1-yl]-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)methanone | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
7EI2
 
 | | Structure of human NNMT in complex with macrocyclic peptide 8 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 8 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7EHZ
 
 | | Structure of human NNMT in complex with macrocyclic peptide 2 | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide 2 | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-30 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
7EGU
 
 | | Structure of human NNMT in complex with macrocyclic peptide X | | Descriptor: | Nicotinamide N-methyltransferase, macrocyclic peptide X | | Authors: | Hayashi, K, Mikamiyama, H, Uehara, S, Yamamoto, S, Cary, D, Nishikawa, J, Ueda, T, Ozasa, H, Mihara, K, Yoshimura, N, Kawai, T, Ono, T, Yamamoto, S, Fumoto, M. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Macrocyclic Peptides as a Novel Class of NNMT Inhibitors: A SAR Study Aimed at Inhibitory Activity in the Cell.
Acs Med.Chem.Lett., 12, 2021
|
|
9IP8
 
 | | Poly-alanine model for HL-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (closed conformation) | | Descriptor: | Epidermal growth factor receptor, HL-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
9IPD
 
 | | Poly-alanine model for LH-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (middle conformation) | | Descriptor: | Epidermal growth factor receptor, LH-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
9IP7
 
 | | Local refinement structure of sEGFR and 528 Fv (from HL-type bispecific diabody Ex3) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 528 Fv from HL-type bispecific diabody Ex3, ... | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
9IPC
 
 | | Poly-alanine model for LH-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (closed conformation) | | Descriptor: | Epidermal growth factor receptor, LH-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
9IPE
 
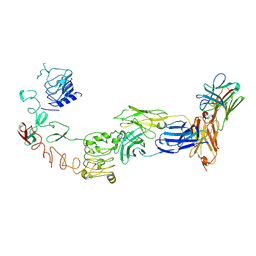 | | Poly-alanine model for LH-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (open conformation) | | Descriptor: | Epidermal growth factor receptor, LH-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
9IP9
 
 | | Poly-alanine model for HL-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (middle conformation) | | Descriptor: | Epidermal growth factor receptor, HL-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
9IPA
 
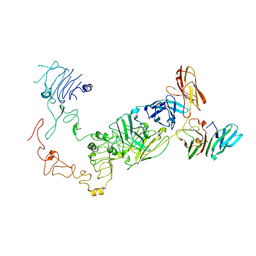 | | Poly-alanine model for HL-type bispecific diabody Ex3 composed of 528 and OKT3 Fvs in ternary complex with sEGFR and CD3gamma-epsilon (open conformation) | | Descriptor: | Epidermal growth factor receptor, HL-type bispecific diabody Ex3, T-cell surface glycoprotein CD3 gamma chain,T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
9IPB
 
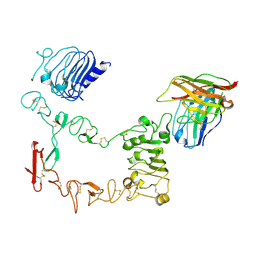 | | Local refinement structure of sEGFR and 528 Fv (from LH-type bispecific diabody Ex3) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 528 Fv from LH-type bispecific diabody Ex3, ... | | Authors: | Sato, K, Uehara, S, Tsugita, A, Matsui, T, Asano, R, Makabe, K, Yokoyama, T, Tanaka, Y. | | Deposit date: | 2024-07-10 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bispecific antibody-antigen complex structures reveal activity enhancement by domain rearrangement
Cell Rep, 2025
|
|
7VU6
 
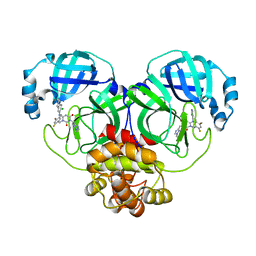 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 3 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Yamamoto, S, Yamane, J, Tachibana, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
7VTH
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with compound 1 | | Descriptor: | 2-[4-[[4-[bis(fluoranyl)methoxy]-2-methyl-phenyl]amino]-2,6-bis(oxidanylidene)-3-[[3,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazin-1-yl]-N-methyl-ethanamide, 3C-like proteinase | | Authors: | Yamamoto, S, Tachibana, Y. | | Deposit date: | 2021-10-29 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19.
J.Med.Chem., 65, 2022
|
|
3WID
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with NADP | | Descriptor: | Glucose 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WIC
 
 | | Structure of a substrate/cofactor-unbound glucose dehydrogenase | | Descriptor: | Glucose 1-dehydrogenase, PENTAETHYLENE GLYCOL, S-1,2-PROPANEDIOL, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WIE
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with D-glucose and NAADP | | Descriptor: | Glucose 1-dehydrogenase, ZINC ION, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
