2QX0
 
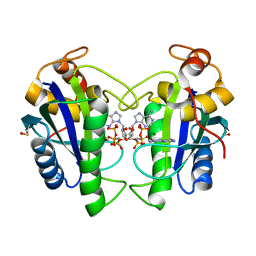 | | Crystal Structure of Yersinia pestis HPPK (Ternary Complex) | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Blaszczyk, J, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2007-08-10 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1LVM
 
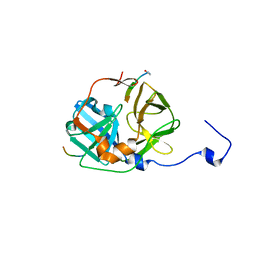 | | CATALYTICALLY ACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH PRODUCT | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
1LVB
 
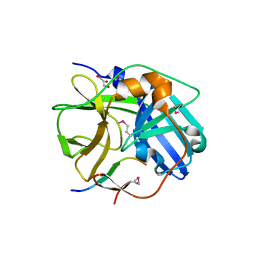 | | CATALYTICALLY INACTIVE TOBACCO ETCH VIRUS PROTEASE COMPLEXED WITH SUBSTRATE | | Descriptor: | CATALYTIC DOMAIN OF THE NUCLEAR INCLUSION PROTEIN A (NIA), GLYCEROL, OLIGOPEPTIDE SUBSTRATE FOR THE PROTEASE | | Authors: | Phan, J, Zdanov, A, Evdokimov, A.G, Tropea, J.E, Peters III, H.K, Kapust, R.B, Li, M, Wlodawer, A, Waugh, D.S. | | Deposit date: | 2002-05-28 | | Release date: | 2002-11-27 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the substrate specificity of tobacco etch virus protease.
J.Biol.Chem., 277, 2002
|
|
6W7L
 
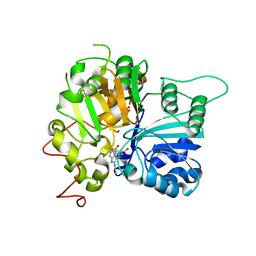 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ632p | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyrazin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
6W7K
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ634p | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyridin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
8SZE
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase in complex with Trimethoprim
To be published
|
|
8SZD
 
 | | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, MAGNESIUM ION, ... | | Authors: | Shaw, G.X, Cherry, S, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of Yersinia pestis dihydrofolate reductase at 1.25-A resolution
To be published
|
|
7UFZ
 
 | | Crystal structure of TDP1 complexed with compound XZ768 | | Descriptor: | (4-{[(4S)-2-phenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)phosphonic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
7UFY
 
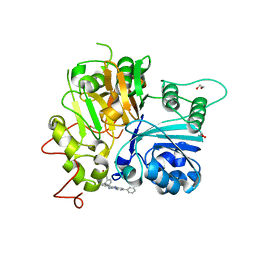 | | Crystal structure of TDP1 complexed with compound XZ766 | | Descriptor: | 1,2-ETHANEDIOL, Tyrosyl-DNA phosphodiesterase 1, [(4-{[(4S)-2,7-diphenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)methyl]phosphonic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
7UPZ
 
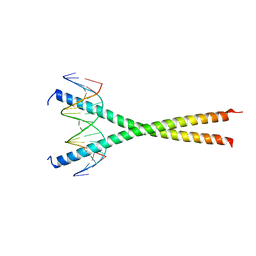 | | Structural basis for cell type specific DNA binding of C/EBPbeta: the case of cell cycle inhibitor p15INK4b promoter | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(*AP*TP*TP*CP*TP*TP*AP*AP*GP*AP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*CP*TP*TP*TP*CP*TP*TP*AP*AP*GP*AP*A)-3') | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Wlodawer, A, Miller, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter.
J.Struct.Biol., 214, 2022
|
|
4M2Z
 
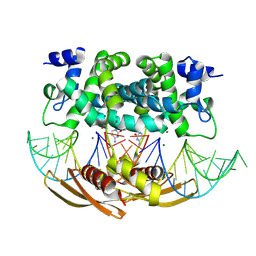 | | Crystal structure of RNASE III complexed with double-stranded RNA and CMP (TYPE II CLEAVAGE) | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA10, ... | | Authors: | Gan, J, Liang, Y.-H, Shaw, G.X, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | RNase III: Genetics and Function; Structure and Mechanism.
Annu. Rev. Genet., 47, 2013
|
|
4M30
 
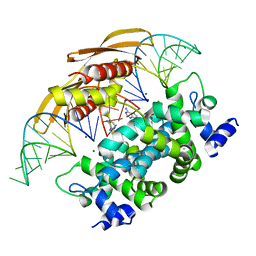 | | Crystal structure of RNASE III complexed with double-stranded RNA AND AMP (TYPE II CLEAVAGE) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gan, J, Liang, Y.-H, Shaw, G.X, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | RNase III: Genetics and Function; Structure and Mechanism.
Annu. Rev. Genet., 47, 2013
|
|
6DIM
 
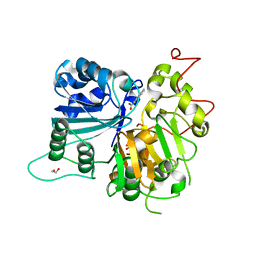 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT1982 from cocktail soak | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DJD
 
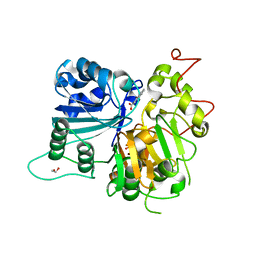 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT1982 (single soak) | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 2019
|
|
6DHU
 
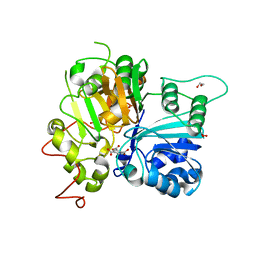 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment ZT0911 from cocktail soak | | Descriptor: | 1,2-ETHANEDIOL, Tyrosyl-DNA phosphodiesterase 1, benzene-1,2,4-tricarboxylic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DIE
 
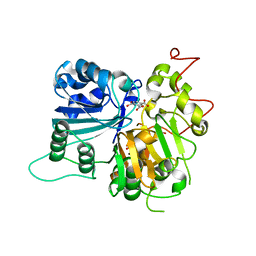 | | Crystal structure of Tdp1 catalytic domain in complex with Zenobia fragment benzene-1,2,4-tricarboxylic acid from single soak | | Descriptor: | 1,2-ETHANEDIOL, Tdp1 catalytic domain (residues 149-609), benzene-1,2,4-tricarboxylic acid | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DJF
 
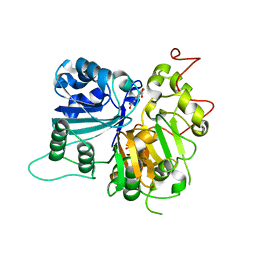 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ502 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxyquinoline-3,8-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
8CVQ
 
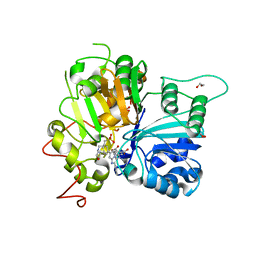 | | Crystal structure of TDP1 complexed with compound XZ761 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-{[(4S)-2,7-diphenylimidazo[1,2-a]pyridin-3-yl]amino}benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of multidentate tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors that simultaneously access the DNA, protein and catalytic-binding sites by oxime diversification.
Rsc Chem Biol, 4, 2023
|
|
8CW2
 
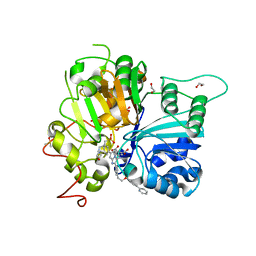 | | Crystal structure of TDP1 complexed with compound XZ760 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-({(4R)-7-phenyl-2-[4-(2-{[4-(pyridin-2-yl)phenyl]methoxy}ethyl)phenyl]imidazo[1,2-a]pyridin-3-yl}amino)benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Wang, W, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.811 Å) | | Cite: | Identification of multidentate tyrosyl-DNA phosphodiesterase 1 (TDP1) inhibitors that simultaneously access the DNA, protein and catalytic-binding sites by oxime diversification.
Rsc Chem Biol, 4, 2023
|
|
8DHJ
 
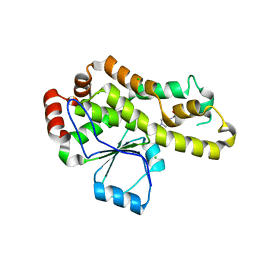 | |
8DHI
 
 | |
5F6D
 
 | | Crystal structure of Ubc9 (K48A/K49A/E54A) complexed with Fragment 6 | | Descriptor: | 6~{H}-benzo[c][1,2]benzothiazine 5,5-dioxide, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-05 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6W
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol) | | Descriptor: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6V
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 1 (biphenol from fragment cocktail screen) | | Descriptor: | 2-(2-hydroxyphenyl)phenol, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5F6X
 
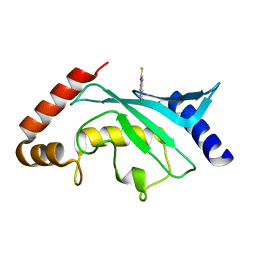 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole from cocktail screen) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
