2ANY
 
 | | Expression, Crystallization and the Three-dimensional Structure of the Catalytic Domain of Human Plasma Kallikrein: Implications for Structure-Based Design of Protease Inhibitors | | Descriptor: | BENZAMIDINE, PHOSPHATE ION, plasma kallikrein, ... | | Authors: | Tang, J, Yu, C.L, Williams, S.R, Springman, E, Jeffery, D, Sprengeler, P.A, Estevez, A, Sampang, J, Shrader, W, Spencer, J.R, Young, W.B, McGrath, M.E, Katz, B.A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expression, crystallization, and three-dimensional structure of the catalytic domain of human plasma kallikrein.
J.Biol.Chem., 280, 2005
|
|
2ANW
 
 | | Expression, crystallization and three-dimensional structure of the catalytic domain of human plasma kallikrein: Implications for structure-based design of protease inhibitors | | Descriptor: | BENZAMIDINE, plasma kallikrein, light chain | | Authors: | Tang, J, Yu, C.L, Williams, S.R, Springman, E, Jeffery, D, Sprengeler, P.A, Estevez, A, Sampang, J, Shrader, W, Spencer, J.R, Young, W.B, McGrath, M.E, Katz, B.A. | | Deposit date: | 2005-08-11 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Expression, crystallization, and three-dimensional structure of the catalytic domain of human plasma kallikrein.
J.Biol.Chem., 280, 2005
|
|
7CBB
 
 | |
3IYM
 
 | | Backbone Trace of the Capsid Protein Dimer of a Fungal Partitivirus from Electron Cryomicroscopy and Homology Modeling | | Descriptor: | Capsid protein | | Authors: | Tang, J, Pan, J, Havens, W.F, Ochoa, W.F, Li, H, Sinkovits, R.S, Guu, T.S.Y, Ghabrial, S.A, Nibert, M.L, Tao, J.Y, Baker, T.S. | | Deposit date: | 2010-02-05 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Backbone Trace of Partitivirus Capsid Protein from Electron Cryomicroscopy and Homology Modeling
Biophys.J., 99, 2010
|
|
3J0F
 
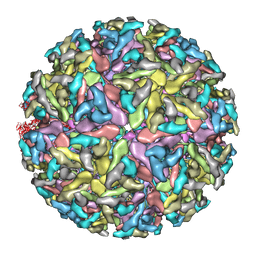 | | Sindbis virion | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Tang, J, Jose, J, Zhang, W, Chipman, P, Kuhn, R.J, Baker, T.S. | | Deposit date: | 2011-07-08 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus.
J.Mol.Biol., 414, 2011
|
|
7NC0
 
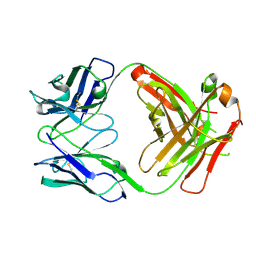 | | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer | | Descriptor: | A33 Fab heavy chain, A33 Fab light chain | | Authors: | Tang, J, Zhang, C, Dalby, P, Kozielski, F. | | Deposit date: | 2021-01-28 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer
To Be Published
|
|
7NFA
 
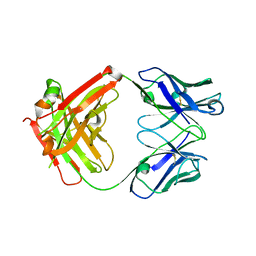 | | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer | | Descriptor: | A33 Fab heavy chain, A33 Fab light chain | | Authors: | Tang, J, Zhang, C, Dalby, P, Kozielski, F. | | Deposit date: | 2021-02-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the humanised A33 Fab C226S variant, an immunotherapy candidate for colorectal cancer
To Be Published
|
|
6M11
 
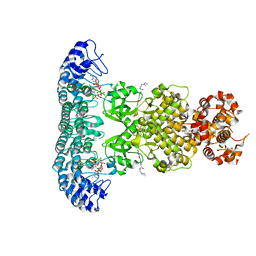 | | Crystal structure of Rnase L in complex with Sunitinib | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M12
 
 | | Crystal Structure of Rnase L in complex with SU11652 | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
6M13
 
 | | Crystal structure of Rnase L in complex with Toceranib | | Descriptor: | 5-[(Z)-(5-fluoranyl-2-oxidanylidene-1H-indol-3-ylidene)methyl]-2,4-dimethyl-N-(2-pyrrolidin-1-ylethyl)-1H-pyrrole-3-carboxamide, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2020-02-24 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
9JDT
 
 | |
9JDQ
 
 | |
6KNK
 
 | | Crystal structure of SbnH in complex with citryl-diaminoethane | | Descriptor: | (2S)-2-{2-[(2-AMINOETHYL)AMINO]-2-OXOETHYL}-2-HYDROXYBUTANEDIOIC ACID, (2~{S})-2-[2-[2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]ethylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, PHOSPHATE ION, ... | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
6KNI
 
 | |
6KNH
 
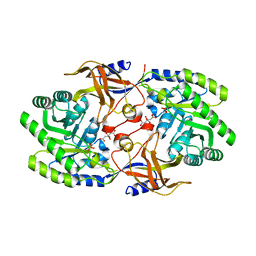 | | Crystal structure of SbnH in complex with citrate, a PLP-dependent decarboxylase in Staphyloferrin B biothesynthesis | | Descriptor: | CITRIC ACID, PHOSPHATE ION, Probable diaminopimelate decarboxylase protein | | Authors: | Tang, J, Ju, Y, Zhou, H. | | Deposit date: | 2019-08-05 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Insights into Substrate Recognition and Activity Regulation of the Key Decarboxylase SbnH in Staphyloferrin B Biosynthesis.
J.Mol.Biol., 431, 2019
|
|
7YIC
 
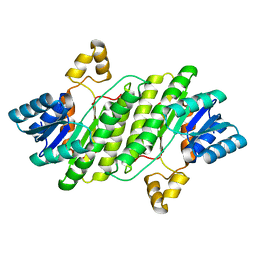 | |
7DSY
 
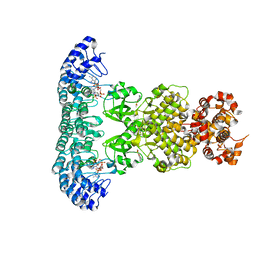 | | Crystal Structure of RNase L in complex with KM05073 | | Descriptor: | 1-chloranyl-3-methylsulfinyl-6,7-dihydro-5H-2-benzothiophen-4-one, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-03 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7DTS
 
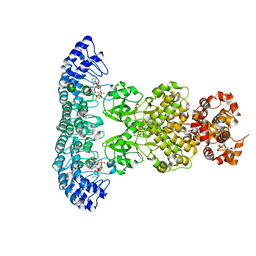 | | Crystal structure of RNase L in complex with AC40357 | | Descriptor: | 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-01-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
7ELW
 
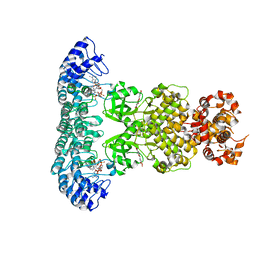 | | Crystal structure of RNase L in complex with Myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, PHOSPHATE ION, Ribonuclease L, ... | | Authors: | Tang, J, Huang, H. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Identification of Small Molecule Inhibitors of RNase L by Fragment-Based Drug Discovery
J.Med.Chem., 65, 2022
|
|
8ZUF
 
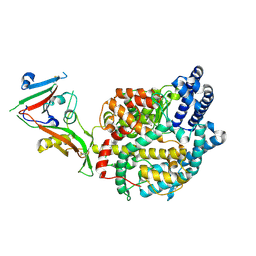 | | Cryo-EM structure of P.nat ACE2 mutant in complex with MOW15-22 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Tang, J, Deng, Z. | | Deposit date: | 2024-06-09 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses.
Cell, 188, 2025
|
|
5GR8
 
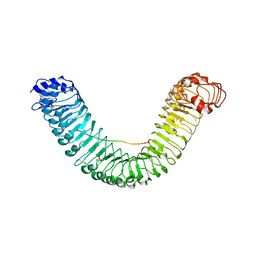 | | Crystal structure of PEPR1-AtPEP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Elicitor peptide 1, ... | | Authors: | Chai, J.J, Tang, J. | | Deposit date: | 2016-08-08 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1
Cell Res., 25, 2015
|
|
9BZG
 
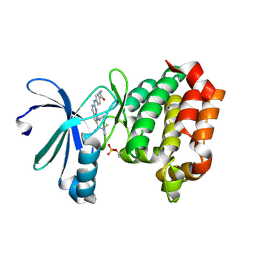 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-[4-(methylcarbamoyl)anilino]-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
9BZL
 
 | | Targeting N-Myc in Neuroblastoma with Selective Aurora Kinase A Degraders | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-{[5-(methylcarbamoyl)pyridin-2-yl]amino}-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A | | Authors: | Baker, Z.D, Tang, J, Shi, K, Aihara, H, Levinson, N.M, Harki, D.A. | | Deposit date: | 2024-05-24 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Targeting N-Myc in neuroblastoma with selective Aurora kinase A degraders.
Cell Chem Biol, 32, 2025
|
|
7E24
 
 | |
7E28
 
 | |
