5XAU
 
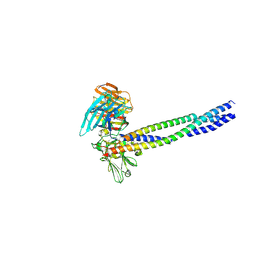 | | Crystal structure of integrin binding fragment of laminin-511 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Takizawa, M, Arimori, T, Kitago, Y, Takagi, J, Sekiguchi, K. | | Deposit date: | 2017-03-15 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic basis for the recognition of laminin-511 by alpha 6 beta 1 integrin.
Sci Adv, 3, 2017
|
|
5MNW
 
 | |
5TPX
 
 | | Bromodomain from Plasmodium Faciparum Gcn5, complexed with compound | | Descriptor: | (1S,2S)-N~1~,N~1~-dimethyl-N~2~-(3-methyl[1,2,4]triazolo[3,4-a]phthalazin-6-yl)-1-phenylpropane-1,2-diamine, CHLORIDE ION, Histone acetyltransferase GCN5, ... | | Authors: | Lin, Y.H, Hou, C.F.D, MOUSTAKIM, M, DIXON, D.J, Loppnau, P, Tempel, W, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, BRENNAN, P.E, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a PCAF Bromodomain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2LKV
 
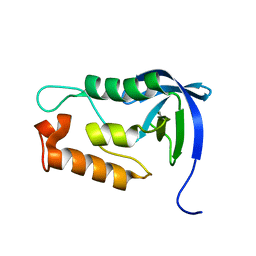 | | Staphylococcal Nuclease PHS variant | | Descriptor: | Thermonuclease | | Authors: | Matzapetakis, M, Pais, T.M, Lamosa, P, Turner, D.L, Santos, H. | | Deposit date: | 2011-10-21 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mannosylglycerate stabilizes staphylococcal nuclease with restriction of slow beta-sheet motions.
Protein Sci., 21, 2012
|
|
6HT0
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with compound 94 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-~{N}-[2-[[(2~{S})-2-methylpyrrolidin-1-yl]methyl]-3~{H}-benzimidazol-5-yl]indazole-5-carboxamide, Protein ENL, ... | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of an MLLT1/3 YEATS Domain Chemical Probe.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
2CWV
 
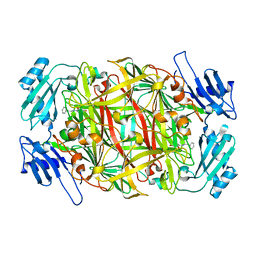 | | Product schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWU
 
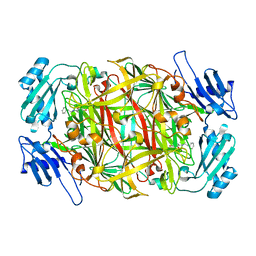 | | Substrate schiff-base intermediate of copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2CWT
 
 | | Catalytic base deletion in copper amine oxidase from arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Chiu, Y.C, Okajima, T, Murakawa, T, Uchida, M, Taki, M, Hirota, S, Kim, M, Yamaguchi, H, Kawano, Y, Kamiya, N, Kuroda, S, Hayashi, H, Yamamoto, Y, Tanizawa, K. | | Deposit date: | 2005-06-26 | | Release date: | 2006-05-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Kinetic and Structural Studies on the Catalytic Role of the Aspartic Acid Residue Conserved in Copper Amine Oxidase(,)
Biochemistry, 45, 2006
|
|
2D1W
 
 | | Substrate Schiff-Base intermediate with tyramine in copper amine oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Kuroda, S, Nakamoto, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2005-09-01 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction
Biochem.Biophys.Res.Commun., 342, 2006
|
|
2E2V
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with benzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2E2T
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with phenylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Kuroda, S, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
2E2U
 
 | | Substrate Schiff-base analogue of copper amine oxidase from Arthrobacter globiformis formed with 4-hydroxybenzylhydrazine | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase | | Authors: | Murakawa, T, Okajima, T, Taki, M, Yamamoto, Y, Hayashi, H, Tanizawa, K. | | Deposit date: | 2006-11-17 | | Release date: | 2007-11-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Catalytic Regulation Conducted by the Substrate Schiff Base and Conserved Aspartic Acid Residue in Bacterial Copper Amine Oxidase Reaction
To be Published
|
|
3W39
 
 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
4MJI
 
 | | T cell response to a HIV reverse transcriptase epitope presented by the protective allele HLA-B*51:01 | | Descriptor: | Beta-2-microglobulin, HIV Reverse Transcriptase peptide Marker, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Motozono, C, Takiguchi, M. | | Deposit date: | 2013-09-03 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Molecular basis of a dominant T cell response to an HIV reverse transcriptase 8-mer epitope presented by the protective allele HLA-B*51:01
J.Immunol., 192, 2014
|
|
7X9U
 
 | | Type-II KH motif of human mitochondrial RbfA | | Descriptor: | Putative ribosome-binding factor A, mitochondrial | | Authors: | Kuwasako, K, Suzuki, S, Furue, M, Takizawa, M, Takahashi, M, Tsuda, K, Nagata, T, Watanabe, S, Tanaka, A, Kobayashi, N, Kigawa, T, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1 H, 13 C, and 15 N resonance assignments and solution structures of the KH domain of human ribosome binding factor A, mtRbfA, involved in mitochondrial ribosome biogenesis.
Biomol.Nmr Assign., 16, 2022
|
|
7VH9
 
 | | Solution structure of the chimeric peptide of the first SURP domain of Human SF3A1 and the interacting region of SF1. | | Descriptor: | Splicing factor 3A subunit 1,Splicing factor 1 | | Authors: | Muto, Y, Kuwasako, K, Takizawa, M, Kobayashi, N, Sakamoto, T. | | Deposit date: | 2021-09-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction between the first SURP domain of the SF3A1 subunit in U2 snRNP and the human splicing factor SF1.
Protein Sci., 31, 2022
|
|
2MPF
 
 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6HPW
 
 | | Crystal structure of ENL (MLLT1) in complex with compound 20 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
1E28
 
 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM2(TAFTIPSI) | | Descriptor: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN, PEPTIDE | | Authors: | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
1E27
 
 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM1(LPPVVAKEI) | | Descriptor: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HIV-1 PEPTIDE (LPPVVAKEI), HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN | | Authors: | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2000-05-18 | | Release date: | 2000-09-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
2ZL8
 
 | |
6OGN
 
 | | Crystal structure of mouse protein arginine methyltransferase 7 in complex with SGC8158 chemical probe | | Descriptor: | 5'-S-(4-{[(4'-chloro[1,1'-biphenyl]-3-yl)methyl]amino}butyl)-5'-thioadenosine, Protein arginine N-methyltransferase 7, UNKNOWN ATOM OR ION, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response.
Nat Commun, 11, 2020
|
|
2N0M
 
 | | The solution structure of the soluble form of the Lipid-modified Azurin from Neisseria gonorrhoeae | | Descriptor: | COPPER (I) ION, Lipid modified azurin protein | | Authors: | Pauleta, S.R, Matzapetakis, M.F, Nobrega, C.F, Carreira, C, Saraiva, I.H. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase.
Biochim.Biophys.Acta, 1857, 2016
|
|
6JNO
 
 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
5HGH
 
 | | HLA*A2402 complexed with HIV nef138 10mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
