1V1S
 
 | |
1V19
 
 | | 2-KETO-3-DEOXYGLUCONATE KINASE FROM THERMUS THERMOPHILUS | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-KETO-3-DEOXYGLUCONATE KINASE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Thermus Thermophilus 2-Keto-3-Deoxygluconate Kinase: Evidence for Recognition of an Open Chain Substrate
J.Mol.Biol., 340, 2004
|
|
1US5
 
 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1US4
 
 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E, Takahashi, H. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1USC
 
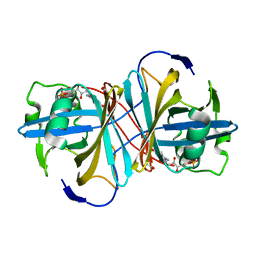 | |
1UZB
 
 | | 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
1USF
 
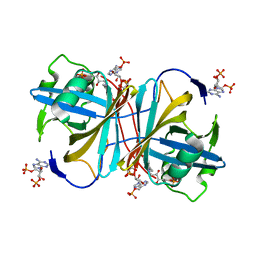 | |
4Y97
 
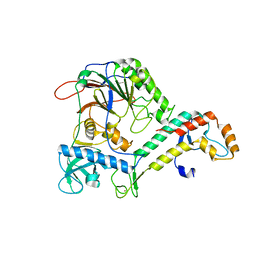 | | Crystal Structure of human Pol alpha B-subunit in complex with C-terminal domain of catalytic subunit | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ZINC ION | | Authors: | Suwa, Y, Gu, J, Baranovskiy, A.G, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of the Human Pol alpha B Subunit in Complex with the C-terminal Domain of the Catalytic Subunit.
J.Biol.Chem., 290, 2015
|
|
3E0J
 
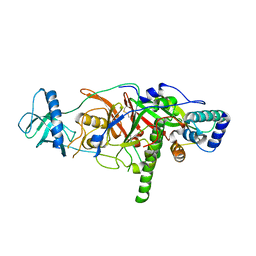 | | X-ray structure of the complex of regulatory subunits of human DNA polymerase delta | | Descriptor: | DNA polymerase subunit delta-2, DNA polymerase subunit delta-3 | | Authors: | Baranovskiy, A.G, Babayeva, N.D, Pavlov, Y.I, Vassylyev, D.G, Tahirov, T.H. | | Deposit date: | 2008-07-31 | | Release date: | 2008-12-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of the complex of regulatory subunits of human DNA polymerase delta.
Cell Cycle, 7, 2008
|
|
2O5I
 
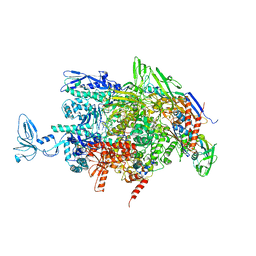 | | Crystal structure of the T. thermophilus RNA polymerase elongation complex | | Descriptor: | 5'-D(*AP*AP*CP*GP*CP*CP*AP*GP*AP*CP*AP*GP*GP*G)-3', 5'-D(P*CP*CP*CP*TP*GP*TP*CP*TP*GP*GP*CP*GP*TP*TP*CP*GP*CP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*GP*AP*GP*UP*CP*UP*GP*CP*GP*GP*CP*GP*CP*GP*CP*G)-3', ... | | Authors: | Vassylyev, D.G, Tahirov, T.H, Vassylyeva, M.N. | | Deposit date: | 2006-12-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for transcription elongation by bacterial RNA polymerase.
Nature, 448, 2007
|
|
5VBN
 
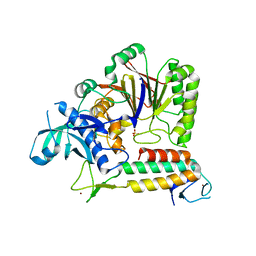 | | Crystal Structure of human DNA polymerase epsilon B-subunit in complex with C-terminal domain of catalytic subunit | | Descriptor: | DNA polymerase epsilon catalytic subunit A, DNA polymerase epsilon subunit 2, SULFATE ION, ... | | Authors: | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2017-03-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human Pol B-subunit in complex with the C-terminal domain of the catalytic subunit.
J. Biol. Chem., 292, 2017
|
|
4QCL
 
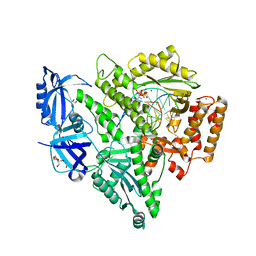 | | Crystal structure of the catalytic core of human DNA polymerase alpha in ternary complex with an RNA-primed DNA template and dCTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA TEMPLATE, ... | | Authors: | Baranovskiy, A.G, Suwa, Y, Babayeva, N.D, Gu, J, Tahirov, T.H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activity and fidelity of human DNA polymerase alpha depend on primer structure
J.Biol.Chem., 2018
|
|
4RR2
 
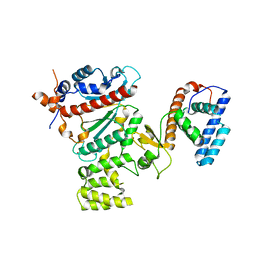 | | Crystal structure of human primase | | Descriptor: | DNA primase large subunit, DNA primase small subunit, IRON/SULFUR CLUSTER, ... | | Authors: | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2014-11-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the human primase.
J.Biol.Chem., 290, 2015
|
|
3MFK
 
 | |
3Q36
 
 | |
3RI4
 
 | |
2IPC
 
 | | Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer | | Descriptor: | Preprotein translocase SecA subunit | | Authors: | Vassylyev, D.G, Mori, H, Vassylyeva, M.N, Tsukazaki, T, Kimura, Y, Tahirov, T.H, Ito, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Translocation ATPase SecA from Thermus thermophilus Reveals a Parallel, Head-to-Head Dimer.
J.Mol.Biol., 364, 2006
|
|
4NYP
 
 | |
4OR5
 
 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4 | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Gu, J, Babayeva, N.D, Suwa, Y, Baranovskiy, A.G, Price, D.H, Tahirov, T.H. | | Deposit date: | 2014-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2014-06-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4.
Cell Cycle, 13, 2014
|
|
4Q5V
 
 | | Crystal structure of the catalytic core of human DNA polymerase alpha in ternary complex with an RNA-primed DNA template and aphidicolin | | Descriptor: | (3R,4R,4aR,6aS,8R,9R,11aS,11bS)-4,9-bis(hydroxymethyl)-4,11b-dimethyltetradecahydro-8,11a-methanocyclohepta[a]naphthale ne-3,9-diol, DNA polymerase alpha catalytic subunit, DNA template, ... | | Authors: | Baranovskiy, A.G, Babayeva, N.D, Suwa, Y, Gu, J, Tahirov, T.H. | | Deposit date: | 2014-04-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural basis for inhibition of DNA replication by aphidicolin.
Nucleic Acids Res., 42, 2014
|
|
1ODE
 
 | |
1OD6
 
 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OI7
 
 | | The Crystal Structure of Succinyl-CoA synthetase alpha subunit from Thermus Thermophilus | | Descriptor: | SUCCINYL-COA SYNTHETASE ALPHA CHAIN | | Authors: | Takahashi, H, Tokunaga, Y, Kuroishi, C, Babayeva, N, Kuramitsu, S, Yokoyama, S, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | The Crystal Structure of Succinyl-Coa Synthetase from Thermus Thermophilus
To be Published
|
|
1J3L
 
 | | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis | | Descriptor: | CHLORIDE ION, Demethylmenaquinone Methyltransferase, MAGNESIUM ION | | Authors: | Rehse, P.H, Miyano, M, Tahirov, T.H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the RNA-processing inhibitor RraA from Thermus thermophilis.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2A6E
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
