8SYO
 
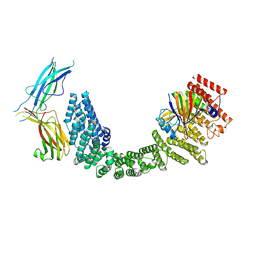 | | Human Retriever VPS35L/VPS29/VPS26C Complex (Composite Map) | | 分子名称: | VPS35 endosomal protein-sorting factor-like, Vacuolar protein sorting-associated protein 26C, Vacuolar protein sorting-associated protein 29 | | 著者 | Chen, Z, Chen, B, Burstein, E, Han, Y. | | 登録日 | 2023-05-25 | | 公開日 | 2023-11-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Structural organization of the retriever-CCC endosomal recycling complex.
Nat.Struct.Mol.Biol., 2023
|
|
1DJ6
 
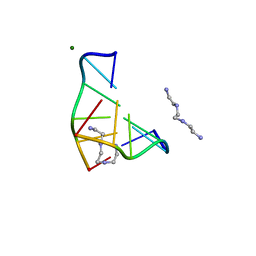 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | 分子名称: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | 著者 | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | 登録日 | 1999-12-01 | | 公開日 | 1999-12-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
3DF0
 
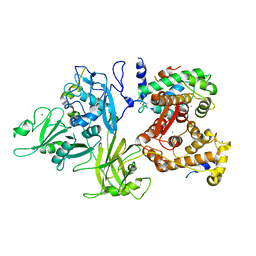 | | Calcium-dependent complex between m-calpain and calpastatin | | 分子名称: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | 著者 | Moldoveanu, T, Gehring, K, Green, D.R. | | 登録日 | 2008-06-11 | | 公開日 | 2008-11-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
7BQS
 
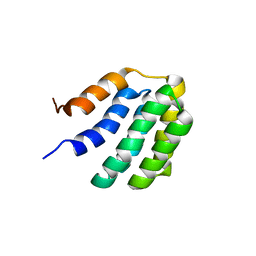 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Nomur | | 著者 | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Mussoc | | 著者 | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
1IT4
 
 | | Solution structure of the prokaryotic Phospholipase A2 from Streptomyces violaceoruber | | 分子名称: | CALCIUM ION, phospholipase A2 | | 著者 | Ohtani, K, Sugiyama, M, Izuhara, M, Koike, T. | | 登録日 | 2002-01-08 | | 公開日 | 2002-09-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.BIOL.CHEM., 277, 2002
|
|
1IT5
 
 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | 分子名称: | Phospholipase A2 | | 著者 | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | 登録日 | 2002-01-09 | | 公開日 | 2002-09-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
7BQN
 
 | | Solution NMR structure of fold-C Rei; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Rei | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQM
 
 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Chantal | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQQ
 
 | | Solution NMR structure of fold-Z Gogy; de novo designed protein with an asymmetric all-alpha topology | | 分子名称: | Gogy | | 著者 | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | 登録日 | 2020-03-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
5JH9
 
 | | Crystal structure of prApe1 | | 分子名称: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | 著者 | Noda, N.N, Adachi, W, Inagaki, F. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JGE
 
 | |
6K3Q
 
 | | Crystal Structure of P450BM3 with N-(3-cyclohexylpropanoyl)-L-prolyl-L-phenylalanine | | 分子名称: | (2S)-2-[[(2S)-1-(3-cyclohexylpropanoyl)pyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | 著者 | Shoji, O, Yonemura, K. | | 登録日 | 2019-05-21 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6L1A
 
 | | Crystal Structure of P450BM3 with N-enanthoyl-L-prolyl-L-phenylalanine | | 分子名称: | (2S)-2-[[(2S)-1-heptanoylpyrrolidin-2-yl]carbonylamino]-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | 著者 | Shoji, O, Yonemura, K. | | 登録日 | 2019-09-28 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Systematic Evolution of Decoy Molecules for the Highly Efficient Hydroxylation of Benzene and Small Alkanes Catalyzed by Wild-Type Cytochrome P450BM3
Acs Catalysis, 10, 2020
|
|
6L1B
 
 | |
7X8Y
 
 | |
7X92
 
 | |
7X90
 
 | |
7X8W
 
 | |
7X8Z
 
 | |
7X91
 
 | |
8J81
 
 | | MDM2 bound with a peptoid | | 分子名称: | (2S)-2-[[(2S)-2-[(6-chloranyl-1H-indol-3-yl)methyl-[(2S)-2-[[(2S)-2-[ethanoyl-(phenylmethyl)amino]propanoyl]-methyl-amino]propanoyl]amino]propanoyl]-methyl-amino]-N-(3,3-dimethylbutyl)-N-[(2S)-1-oxidanylidene-1-piperazin-1-yl-propan-2-yl]propanamide, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Yokomine, M, Fukuda, Y, Ago, H, Matsuura, H, Ueno, G, Nagatoishi, S, Yamamoto, M, Tsumoto, K, Jumpei, M, Sando, S. | | 登録日 | 2023-04-28 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | A structural and physicochemical study of how a peptoid binds to a protein
To Be Published
|
|
4X68
 
 | | Crystal Structure of OP0595 complexed with AmpC | | 分子名称: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, NICKEL (II) ION | | 著者 | Yamada, M, Watanabe, T. | | 登録日 | 2014-12-07 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
6L0V
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NGR2, ... | | 著者 | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | 登録日 | 2019-09-27 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.347 Å) | | 主引用文献 | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
4X69
 
 | | Crystal structure of OP0595 complexed with CTX-M-44 | | 分子名称: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase Toho-1 | | 著者 | Yamada, M, Watanabe, T. | | 登録日 | 2014-12-07 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
