5JGF
 
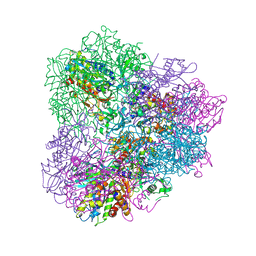 | | Crystal structure of mApe1 | | Descriptor: | Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JHC
 
 | |
7VM1
 
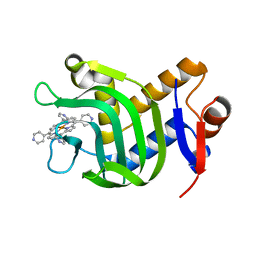 | | Crystal Structure of HasAp Capturing Iron Tetra(4-pyridyl)porphyrin | | Descriptor: | Fe-Tetra(4-pyridyl)porphyrin, GLYCEROL, Heme acquisition protein HasAp | | Authors: | Shisaka, Y, Ueda, G, Sakakibara, E, Sugimoto, H, Shoji, O. | | Deposit date: | 2021-10-06 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tetraphenylporphyrin Enters the Ring: First Example of a Complex between Highly Bulky Porphyrins and a Protein.
Chembiochem, 23, 2022
|
|
1QTO
 
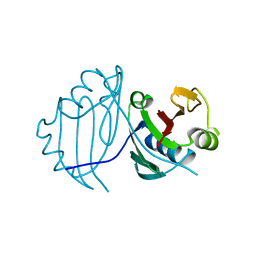 | | 1.5 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT FROM BLEOMYCIN-PRODUCING STREPTOMYCES VERTICILLUS | | Descriptor: | BLEOMYCIN-BINDING PROTEIN | | Authors: | Kawano, Y, Kumagai, T, Muta, K, Matoba, Y, Davies, J, Sugiyama, M. | | Deposit date: | 1999-06-28 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5 A crystal structure of a bleomycin resistance determinant from bleomycin-producing Streptomyces verticillus.
J.Mol.Biol., 295, 2000
|
|
1DJ6
 
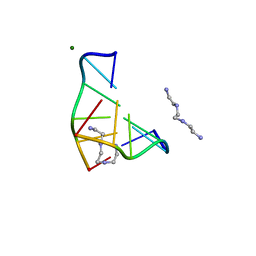 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
1IT4
 
 | | Solution structure of the prokaryotic Phospholipase A2 from Streptomyces violaceoruber | | Descriptor: | CALCIUM ION, phospholipase A2 | | Authors: | Ohtani, K, Sugiyama, M, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-08 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.BIOL.CHEM., 277, 2002
|
|
1IT5
 
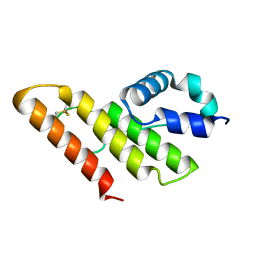 | | Solution structure of apo-type PLA2 from Streptomyces violaceruber A-2688. | | Descriptor: | Phospholipase A2 | | Authors: | Sugiyama, M, Ohtani, K, Izuhara, M, Koike, T. | | Deposit date: | 2002-01-09 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel prokaryotic phospholipase A2. Characterization, gene cloning, and solution structure.
J.Biol.Chem., 277, 2002
|
|
7X8Y
 
 | |
7X8W
 
 | |
7X8Z
 
 | |
7X90
 
 | |
7X92
 
 | |
7X91
 
 | |
7X95
 
 | |
7X96
 
 | |
7X93
 
 | |
7X94
 
 | |
7Y6L
 
 | |
7Y6N
 
 | |
1UEA
 
 | | MMP-3/TIMP-1 COMPLEX | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-3, TISSUE INHIBITOR OF METALLOPROTEINASE-1, ... | | Authors: | Bode, W, Maskos, K, Gomis-Rueth, F.-X, Nagase, H. | | Deposit date: | 1997-06-06 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1.
Nature, 389, 1997
|
|
7JTP
 
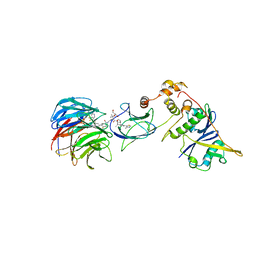 | | Crystal structure of Protac MS67 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | Elongin-B, Elongin-C, GLYCEROL, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
7JTO
 
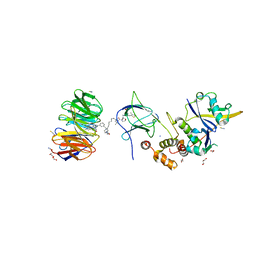 | | Crystal structure of Protac MS33 in complex with the WD repeat-containing protein 5 and pVHL:ElonginC:ElonginB | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(11-{[2-(4-{[4'-(4-methylpiperazin-1-yl)-3'-{[6-oxo-4-(trifluoromethyl)-5,6-dihydropyridine-3-carbonyl]amino}[1,1'-biphenyl]-3-yl]methyl}piperazin-1-yl)ethyl]amino}-11-oxoundecanoyl)-L-valyl-(4R)-4-hydroxy-N-{[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl}-L-prolinamide, Elongin-B, ... | | Authors: | Kottur, J, Jain, R, Aggarwal, A.K. | | Deposit date: | 2020-08-18 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models.
Sci Transl Med, 13, 2021
|
|
4X69
 
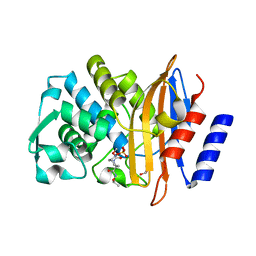 | | Crystal structure of OP0595 complexed with CTX-M-44 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase Toho-1 | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
5ZOH
 
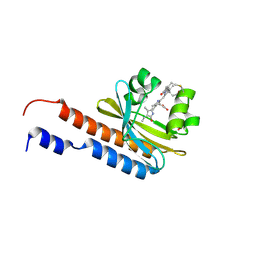 | |
6A6S
 
 | | Crystal structure of the modified fructosyl peptide oxidase from Aspergillus nidulans in complex with FSA, Seleno-methionine Derivative | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, 1-S-(carboxymethyl)-1-thio-beta-D-fructopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ogawa, N, Maruyama, Y, Itoh, T, Hashimoto, W, Murata, K. | | Deposit date: | 2018-06-29 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Creation of haemoglobin A1c direct oxidase from fructosyl peptide oxidase by combined structure-based site specific mutagenesis and random mutagenesis.
Sci Rep, 9, 2019
|
|
