3BAC
 
 | |
3BA8
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 2-amino-7-fluoro-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BAB
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-(4-pyrimidin-2-ylpiperazin-1-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BAA
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-{[2-(1H-imidazol-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BA9
 
 | | Structural Basis for Inhbition of NAD-Dependent Ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, GLYCEROL, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhbition of NAD-Dependent Ligase
To be Published
|
|
3LH5
 
 | |
4L5B
 
 | | Human dCK C4S-S74E mutant in complex with UDP and the DI-43 inhibitor | | Descriptor: | 1-[5-(4-{[(4,6-diaminopyrimidin-2-yl)sulfanyl]methyl}-5-propyl-1,3-thiazol-2-yl)-2-methoxyphenoxy]-2-methylpropan-2-ol, Deoxycytidine kinase, URIDINE-5'-DIPHOSPHATE | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-06-10 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Development of new deoxycytidine kinase inhibitors and noninvasive in vivo evaluation using positron emission tomography.
J.Med.Chem., 56, 2013
|
|
4O3P
 
 | | Crystal structure of human polymerase eta inserting dctp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4O0F
 
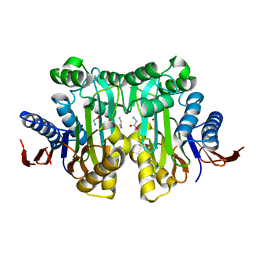 | | Crystal structure of the human L-asparaginase protein T219A mutant | | Descriptor: | GLYCINE, Isoaspartyl peptidase/L-asparaginase, SODIUM ION | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
J.Mol.Biol., 426, 2014
|
|
4O0H
 
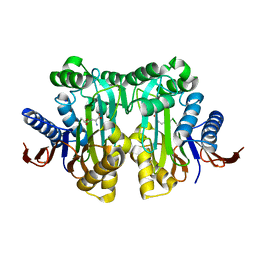 | |
4O3Q
 
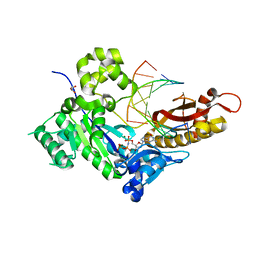 | | Crystal structure of human polymerase eta inserting dgtp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4O0D
 
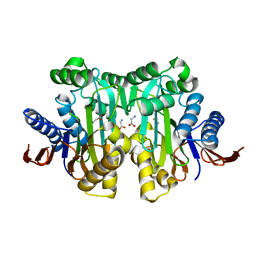 | | Crystal structure of the human L-asparaginase protein T168S mutant | | Descriptor: | GLYCINE, Isoaspartyl peptidase/L-asparaginase, SODIUM ION | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
J.Mol.Biol., 426, 2014
|
|
4O3N
 
 | | Crystal structure of human dna polymerase eta in ternary complex with native dna and incoming nucleotide (dcp) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4O3R
 
 | | Crystal structure of human polymerase eta extending an 8-oxog dna lesion: post insertion of 8-oxog-da pair | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), DNA (5'-D(*CP*AP*TP*GP*(8OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4O0E
 
 | | Crystal structure of the human L-asparaginase protein T186V mutant | | Descriptor: | Isoaspartyl peptidase/L-asparaginase, SODIUM ION | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
J.Mol.Biol., 426, 2014
|
|
4O0C
 
 | | High resolution crystal structure of uncleaved human L-asparaginase protein | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Isoaspartyl peptidase/L-asparaginase, ... | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
J.Mol.Biol., 426, 2014
|
|
4O60
 
 | | Structure of ankyrin repeat protein | | Descriptor: | ANK-N5C-317 | | Authors: | Ethayathulla, A.S, Guan, L. | | Deposit date: | 2013-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A transcription blocker isolated from a designed repeat protein combinatorial library by in vivo functional screen.
Sci Rep, 5, 2015
|
|
4O3O
 
 | | Crystal structure of human polymerase eta inserting datp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4O0G
 
 | | Crystal structure of the human L-asparaginase protein T219V mutant | | Descriptor: | Isoaspartyl peptidase/L-asparaginase, SODIUM ION | | Authors: | Nomme, J, Lavie, A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
J.Mol.Biol., 426, 2014
|
|
4O3S
 
 | | Crystal structure of human polymerase eta extending an 8-oxog dna lesion: post insertion of 8-oxog-dc pair | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(8OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4PV3
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, SODIUM ION | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PU6
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ cations | | Descriptor: | ASPARTIC ACID, L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PV2
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ and Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, NITRATE ION, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Clavel, D, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PVR
 
 | |
4PVS
 
 | |
