7P16
 
 | |
7P14
 
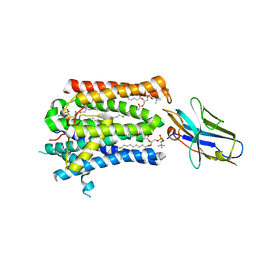 | | Structure of full-length rXKR9 in complex with a sybody at 3.66A | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sybody, ... | | Authors: | Straub, M.S, Sawicka, M, Dutzler, R. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of the caspase activated protein XKR9 involved in apoptotic lipid scrambling.
Elife, 10, 2021
|
|
7P5M
 
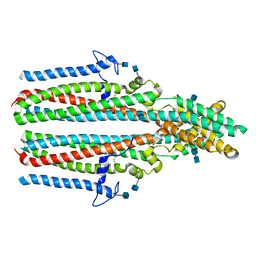 | | Cryo-EM structure of human TTYH2 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5C
 
 | | Cryo-EM structure of human TTYH3 in Ca2+ and GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 3 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7P5J
 
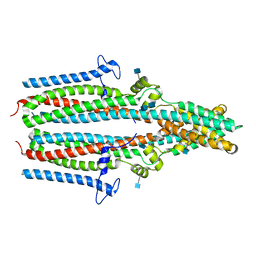 | | Cryo-EM structure of human TTYH1 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 1 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
7QIA
 
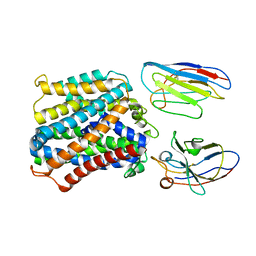 | | Structure of apo-EleNRMT in complex with two nanobodies at 3.5A | | Descriptor: | Divalent metal cation transporter, Nanobody 1, Nanobody 2 | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
7QIC
 
 | | Structure of magnesium-bound EleNRMT in complex with two nanobodies at 4.1A | | Descriptor: | Divalent metal cation transporter, MAGNESIUM ION, Nanobody 1, ... | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-14 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
7QJI
 
 | | X-Ray Structure of apo-EleNRMT in complex with two Nanobodies at 4.1A | | Descriptor: | Divalent metal cation transporter, Elen-Nanobody-complex | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
7QJJ
 
 | | X-Ray Structure of a Mn2+ soak of EleNRMT in complex with two Nanobodies at 4.6A | | Descriptor: | Divalent metal cation transporter, Elen-Nb1-Nb2, MANGANESE (II) ION | | Authors: | Ramanadane, K, Straub, M.S, Dutzler, R, Manatschal, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural and functional properties of a magnesium transporter of the SLC11/NRAMP family.
Elife, 11, 2022
|
|
8ONT
 
 | | Structure of Setaria italica NRAT in complex with a nanobody | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, NRAMP related aluminium transporter, Nanobody1 | | Authors: | Ramanadane, K, Liziczai, M, Markovic, D, Straub, M.S, Rosalen, G.T, Udovcic, A, Dutzler, R, Manatschal, C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural and functional properties of a plant NRAMP-related aluminum transporter.
Elife, 12, 2023
|
|
7P54
 
 | | Cryo-EM structure of human TTYH2 in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein tweety homolog 2 | | Authors: | Sukalskaia, A, Straub, M.S, Sawicka, M, Deneka, D, Dutzler, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions.
Nat Commun, 12, 2021
|
|
8B8G
 
 | | Cryo-EM structure of Ca2+-free mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6 | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8K
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin closed/closed | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8J
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518H mutant in Digitonin | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC1
 
 | | Cryo-EM Structure of Ca2+-bound mTMEM16F F518A_Q623A mutant in GDN | | Descriptor: | Anoctamin-6,mTMEM16F, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8M
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F N562A mutant in Digitonin open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8B8Q
 
 | | Structure of mTMEM16F in lipid Nanodiscs in the presence of Ca2+ | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M.S, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
8BC0
 
 | | Cryo-EM structure of Ca2+-bound mTMEM16F F518A Q623A mutant in GDN open/closed | | Descriptor: | Anoctamin-6, CALCIUM ION | | Authors: | Arndt, M, Alvadia, C, Straub, M, Clerico-Mosina, V, Paulino, C, Dutzler, R. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the activation of the lipid scramblase TMEM16F.
Nat Commun, 13, 2022
|
|
