4CD3
 
 | | RnNTPDase2 X4 variant in complex with PSB-071 | | Descriptor: | 1-AMINO-4-(3-METHYLPHENYL)AMINO-9,10-DIOXO-9,10-DIHYDROANTHRACENE-2-SULFONATE, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE 2, GLYCEROL | | Authors: | Zebisch, M, Schaefer, P, Straeter, N. | | Deposit date: | 2013-10-29 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Ntpdase2 in Complex with the Sulfoanthraquinone Inhibitor Psb-071.
J.Struct.Biol., 185, 2014
|
|
4CG1
 
 | | Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca | | Descriptor: | CUTINASE, SULFATE ION | | Authors: | Roth, C, Wei, R, Oeser, T, Then, J, Foellner, C, Zimmermann, W, Straeter, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Studies on a Thermostable Polyethylene Terephthalate Degrading Hydrolase from Thermobifida Fusca.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4CG2
 
 | | Structural and functional studies on a thermostable polyethylene terephthalate degrading hydrolase from Thermobifida fusca | | Descriptor: | CUTINASE, SULFATE ION, phenylmethanesulfonic acid | | Authors: | Roth, C, Wei, R, Oeser, T, Then, J, Foellner, C, Zimmermann, W, Straeter, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.437 Å) | | Cite: | Structural and Functional Studies on a Thermostable Polyethylene Terephthalate Degrading Hydrolase from Thermobifida Fusca.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4EZY
 
 | |
4EZN
 
 | |
4EZX
 
 | |
4HYB
 
 | |
4HY9
 
 | |
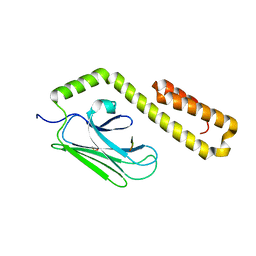
4JWD
 
 | |
4H2B
 
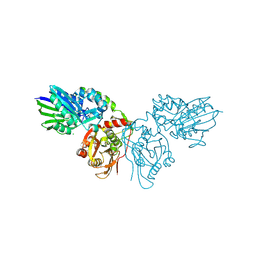 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with Baicalin | | Descriptor: | 5'-nucleotidase, 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, CALCIUM ION, ... | | Authors: | Zebisch, M, Pippel, J, Knapp, K, Straeter, N. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
4H2G
 
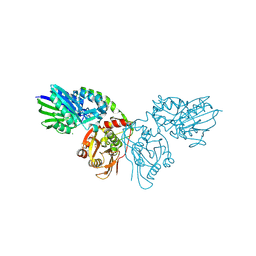 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with adenosine | | Descriptor: | 5'-nucleotidase, ADENOSINE, CALCIUM ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
4H1Y
 
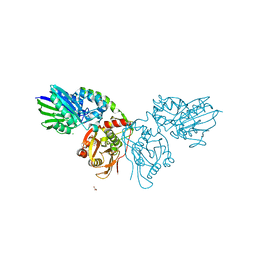 | | Human ecto-5'-nucleotidase (CD73): crystal form II (open) in complex with PSB11552 | | Descriptor: | 2,2'-[(2-{[2-({[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)ethyl]amino}-2-oxoethyl)imino]diacetic acid (non-preferred name), 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Pippel, J, Zebisch, M, Knapp, K, Straeter, N. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
4H2F
 
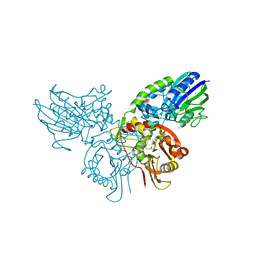 | | Human ecto-5'-nucleotidase (CD73): crystal form I (open) in complex with adenosine | | Descriptor: | 5'-nucleotidase, ADENOSINE, CALCIUM ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
4H2I
 
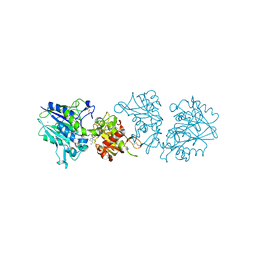 | | Human ecto-5'-nucleotidase (CD73): crystal form III (closed) in complex with AMPCP | | Descriptor: | 5'-nucleotidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Straeter, N, Knapp, K.M, Zebisch, M, Pippel, J. | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-28 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human Ecto-5'-Nucleotidase (CD73): Insights into the Regulation of Purinergic Signaling.
Structure, 20, 2012
|
|
1UTE
 
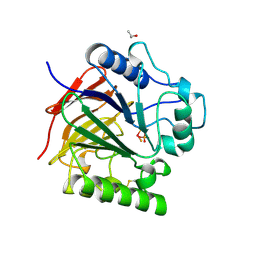 | | PIG PURPLE ACID PHOSPHATASE COMPLEXED WITH PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ISOPROPYL ALCOHOL, MU-OXO-DIIRON, ... | | Authors: | Guddat, L.W, Mcalpine, A, Hume, D, Hamilton, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1999-01-18 | | Release date: | 1999-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of mammalian purple acid phosphatase.
Structure Fold.Des., 7, 1999
|
|
3ZNJ
 
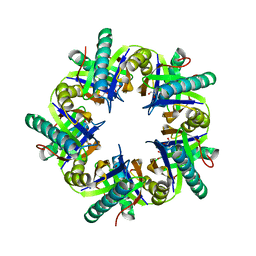 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3ZNU
 
 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
4OT7
 
 | | X-structure of a variant of NCR from zymomonas mobilis | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase/NADH oxidase | | Authors: | Genz, M, Strater, N. | | Deposit date: | 2014-02-13 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Surface Loops Representing Enzyme Modifying Element in the Field of Tuning Enzyme Properties
TO BE PUBLISHED
|
|
