5CXZ
 
 | | SYK catalytic domain complexed with naphthyridine inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, N-{7-[4-(dimethylamino)phenyl]-1,6-naphthyridin-5-yl}propane-1,3-diamine, ... | | Authors: | Thoma, G, Veenstra, S, Strang, R, Blanz, J, Vangrevelinghe, E, Berghausen, J, Lee, C.C, Zerwes, H.-G. | | Deposit date: | 2015-07-29 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Orally bioavailable Syk inhibitors with activity in a rat PK/PD model.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4Y8E
 
 | | PA3825-EAL Ca-Apo Structure | | Descriptor: | CALCIUM ION, PA3825 EAL | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2015-02-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of PA3825 from P. aeruginosa bound to cyclic di-GMP and pGpG: new insights for a potential three-metal catalytic mechanism of EAL domains
To Be Published
|
|
4Y9N
 
 | | PA3825-EAL Metal-Free-Apo Structure - Magnesium Co-crystallisation | | Descriptor: | PA3825-EAL, PHOSPHATE ION | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of PA3825 from P. aeruginosa bound to cyclic di-GMP and pGpG: new insights for a potential three-metal catalytic mechanism of EAL domains
To Be Published
|
|
4Y9O
 
 | | PA3825-EAL Metal-Free-Apo Structure - Manganese Co-crystallisation | | Descriptor: | PA3825-EAL, PHOSPHATE ION | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | MucR and PA3825 EAL-phosphodiesterase domains from Pseudomonas aeruginosa suggest roles for three metals in the active site
To Be Published
|
|
1OZT
 
 | | Crystal Structure of apo-H46R Familial ALS Mutant human Cu,Zn Superoxide Dismutase (CuZnSOD) to 2.5A resolution | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
1OZU
 
 | | Crystal Structure of Familial ALS Mutant S134N of human Cu,Zn Superoxide Dismutase (CuZnSOD) to 1.3A resolution | | Descriptor: | SULFATE ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Elam, J.S, Taylor, A.B, Strange, R, Antonyuk, S, Doucette, P.A, Rodriguez, J.A, Hasnain, S.S, Hayward, L.J, Valentine, J.S, Yeates, T.O, Hart, P.J. | | Deposit date: | 2003-04-09 | | Release date: | 2003-05-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Amyloid-like Filaments and Water-filled Nanotubes Formed by SOD1 Mutant Proteins Linked to Familial ALS
Nat.Struct.Biol., 10, 2003
|
|
5NGX
 
 | | The 1.06 A resolution structure of the L16G mutant of ferric cytochrome c prime from Alcaligenes xylosoxidans, complexed with nitrite | | Descriptor: | Cytochrome c', GLYCEROL, HEME C, ... | | Authors: | Strange, R, Hough, M, Kekelli, D, Horrell, S, Moreno Chicano, T. | | Deposit date: | 2017-03-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Distinguishing Nitro vs Nitrito Coordination in Cytochrome c' Using Vibrational Spectroscopy and Density Functional Theory.
Inorg.Chem., 56, 2017
|
|
5NC0
 
 | | The 0.91 A resolution structure of the L16G mutant of cytochrome c prime from Alcaligenes xylosoxidans, complexed with nitric oxide | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c', HEME C, ... | | Authors: | Strange, R, Hough, M, Antonyuk, S, Rustage, N. | | Deposit date: | 2017-03-02 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Distinguishing Nitro vs Nitrito Coordination in Cytochrome c' Using Vibrational Spectroscopy and Density Functional Theory.
Inorg Chem, 56, 2017
|
|
8PFP
 
 | |
8PFL
 
 | | Crystal structure of WRN helicase domain in complex with 3 | | Descriptor: | 2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-7-oxidanylidene-6-[4-(3-oxidanylpyridin-2-yl)carbonylpiperazin-1-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]-~{N}-[2-methyl-4-(trifluoromethyl)phenyl]ethanamide, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
8PFO
 
 | | Crystal structure of WRN helicase domain in complex with HRO761 | | Descriptor: | Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION, ~{N}-[2-chloranyl-4-(trifluoromethyl)phenyl]-2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-6-[4-(6-methyl-5-oxidanyl-pyrimidin-4-yl)carbonylpiperazin-1-yl]-7-oxidanylidene-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]ethanamide | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
6Q7A
 
 | |
6Q6O
 
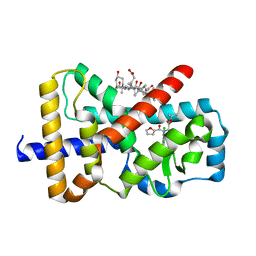 | |
6Q7H
 
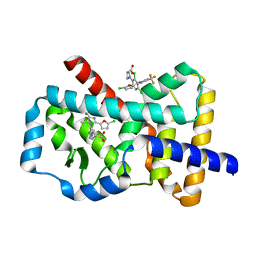 | |
5CY3
 
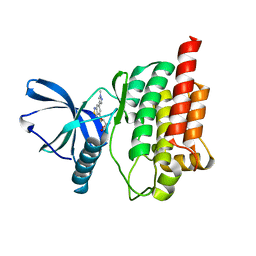 | |
6Q6M
 
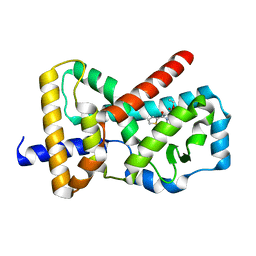 | |
5CXH
 
 | |
4Y9P
 
 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, PA3825-EAL | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | MucR and PA3825 EAL-phosphodiesterase domains from Pseudomonas aeruginosa suggest roles for three metals in the active site
To Be Published
|
|
4Y9M
 
 | | PA3825-EAL Metal-Free-Apo Structure | | Descriptor: | PA3825-EAL, PHOSPHATE ION | | Authors: | Bellini, D, Horrell, S, Wagner, A, Strange, R, Walsh, M.A. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
2WKO
 
 | | Structure of metal loaded Pathogenic SOD1 Mutant G93A. | | Descriptor: | COPPER (II) ION, IODIDE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Antonyuk, S.V, Galaleldeen, A, Strange, R, Whitson, L, Narayana, N, Taylor, A, Schuermann, J.P, Holloway, S.P, Hasnain, S.S, Hart, P.J. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Biophysical Properties of Metal-Free Pathogenic Sod1 Mutants A4V and G93A.
Arch.Biochem.Biophys., 492, 2009
|
|
5MKG
 
 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Diguanylate phosphodiesterase | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
5MFU
 
 | | PA3825-EAL Mn-pGpG Structure | | Descriptor: | Diguanylate phosphodiesterase, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-11-18 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
5N8H
 
 | | Serial Cu nitrite reductase structures at elevated cryogenic temperature, 240K. Dataset 3. | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Horrell, S, Kekilli, D, Hough, M, Strange, R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-site protein dynamics and solvent accessibility in native Achromobacter cycloclastes copper nitrite reductase.
IUCrJ, 4, 2017
|
|
5N8G
 
 | | Serial Cu nitrite reductase structures at elevated cryogenic temperature, 240K. Dataset 2. | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Horrell, S, Kekilli, D, Hough, M, Strange, R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Active-site protein dynamics and solvent accessibility in native Achromobacter cycloclastes copper nitrite reductase.
IUCrJ, 4, 2017
|
|
5N8I
 
 | | Serial Cu nitrite reductase structures at elevated cryogenic temperature, 100K reference dataset. | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION | | Authors: | Horrell, S, Kekilli, D, Hough, M, Strange, R. | | Deposit date: | 2017-02-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Active-site protein dynamics and solvent accessibility in native Achromobacter cycloclastes copper nitrite reductase.
IUCrJ, 4, 2017
|
|
