1E18
 
 | | TUNGSTEN-SUSBSTITUTED DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE., ETHANOL, ... | | Authors: | Bailey, S, Stewart, L.J. | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimethylsulfoxide Reductase: An Enzyme Capable of Catalysis with Either Molybdenum or Tungsten at the Active Site
J.Mol.Biol., 299, 2000
|
|
5DG5
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET IN COMPLEX WITH ALTIRATINIB ANALOG DP-4157 | | Descriptor: | Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxam ide | | Authors: | Smith, B.D, Kaufman, M.D, Leary, C.B, Turner, B.A, Wise, S.A, Ahn, Y.M, Booth, R.J, Caldwell, T.M, Ensinger, C.L, Hood, M.M, Lu, W.-P, Patt, T.W, Patt, W.C, Rutkoski, T.J, Samarakoon, T, Telikepalli, H, Vogeti, L, Vogeti, S, Yates, K.M, Chun, L, Stewart, L.J, Clare, M, Flynn, D.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2.
Mol.Cancer Ther., 14, 2015
|
|
1K4T
 
 | | HUMAN DNA TOPOISOMERASE I (70 KDA) IN COMPLEX WITH THE POISON TOPOTECAN AND COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Staker, B.L, Hjerrild, K, Feese, M.D, Behnke, C.A, Burgin Jr, A.B, Stewart, L.J. | | Deposit date: | 2001-10-08 | | Release date: | 2002-12-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of topoisomerase I poisoning by a camptothecin analog
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K4S
 
 | | HUMAN DNA TOPOISOMERASE I IN COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | 5'-D(*(SPT)P*GP*AP*AP*AP*AP*AP*(5IU)P*(5IU)P*(5IU)P*(5IU)P*T)-3', 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*(5IU)P*(5IU))-3', 5'-D(*AP*AP*AP*AP*AP*TP*(IDO)UP*(IDO)UP*(IDO)UP*(IDO)UP*CP*AP*AP*AP*GP*(IDO)UP*CP*(IDO)UP*(IDO)UP*(IDO)UP*(IDO)UP*T)-3', ... | | Authors: | Staker, B.L, Hjerrild, K, Feese, M.D, Behnke, C.A, Burgin Jr, A.B, Stewart, L.J. | | Deposit date: | 2001-10-08 | | Release date: | 2002-12-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The mechanism of topoisomerase I poisoning by a camptothecin analog
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3QRJ
 
 | | The crystal structure of human abl1 kinase domain T315I mutant in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
3QRK
 
 | | The crystal structure of human abl1 kinase domain in complex with DP-987 | | Descriptor: | (3S)-6-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-1,2,3,4-tetrahydroisoquinoline-3-carboxylic acid, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
3QRI
 
 | | The crystal structure of human abl1 kinase domain in complex with DCC-2036 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, SODIUM ION, Tyrosine-protein kinase ABL1 | | Authors: | Chan, W.W, Wise, S.C, Kaufman, M.D, Ahn, Y.M, Ensinger, C.L, Haack, T, Hood, M.M, Jones, J, Lord, J.W, Lu, W.P, Miller, D, Patt, W.C, Smith, B.D, Petillo, P.A, Rutkoski, T.J, Telikepalli, H, Vogeti, L, Yao, T, Chun, L, Clark, R, Evangelista, P, Gavrilescu, L.C, Lazarides, K, Zaleskas, V.M, Stewart, L.J, Van Etten, R.A, Flynn, D.L. | | Deposit date: | 2011-02-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Control Inhibition of the BCR-ABL1 Tyrosine Kinase, Including the Gatekeeper T315I Mutant, by the Switch-Control Inhibitor DCC-2036.
Cancer Cell, 19, 2011
|
|
3IFT
 
 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
3EIY
 
 | |
6D1N
 
 | |
4IJN
 
 | |
3EOM
 
 | |
3EK2
 
 | |
3EJ2
 
 | |
3EON
 
 | |
3EZO
 
 | |
3DMS
 
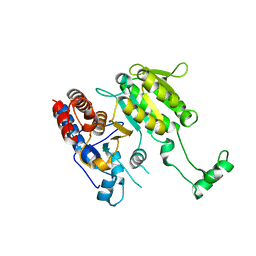 | |
3EJ0
 
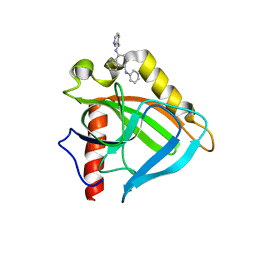 | |
7KUW
 
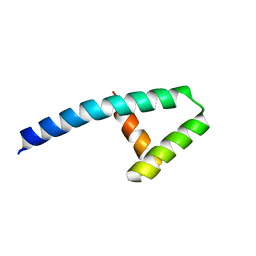 | |
7LMX
 
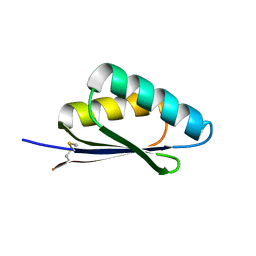 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
4O2D
 
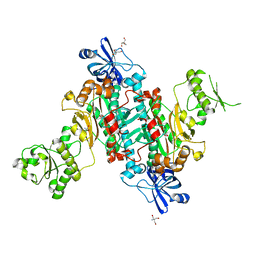 | |
6D41
 
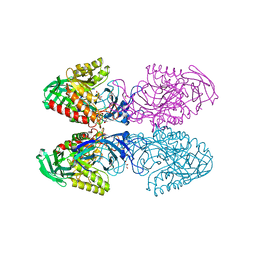 | | Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D50
 
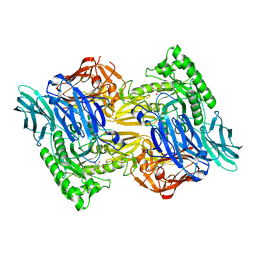 | | Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, CALCIUM ION, Glycosyl hydrolases family 2, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D89
 
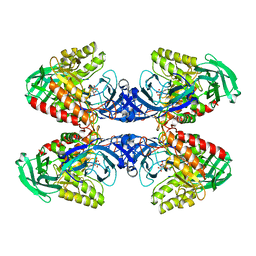 | |
