6U1S
 
 | | Cryo-EM structure of a de novo designed 16-helix transmembrane nanopore, TMHC8_R. | | Descriptor: | de novo designed 16-helix transmembrane nanopore, TMHC8_R | | Authors: | Johnson, M.J, Reggiano, G, Xu, C, Lu, P, Hsia, Y, Brunette, T.J, DiMaio, F, Baker, D, Kollman, J. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Computational Design of Transmembrane Channels
To Be Published
|
|
6M6Z
 
 | | A de novo designed transmembrane nanopore, TMH4C4 | | Descriptor: | TMH4C4 | | Authors: | Lu, P, Xu, C, Reggiano, G, Xu, Q, DiMaio, F, Baker, D. | | Deposit date: | 2020-03-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
7JZL
 
 | |
7JZN
 
 | |
3NNU
 
 | | Crystal structure of P38 alpha in complex with DP1376 | | Descriptor: | 2-{3-[(5E)-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-3-thiophen-2-yl-2,5-dihydro-1H-pyrazol-1-yl]phenyl}acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNW
 
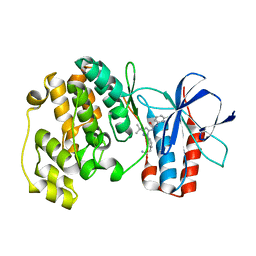 | | Crystal structure of P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNX
 
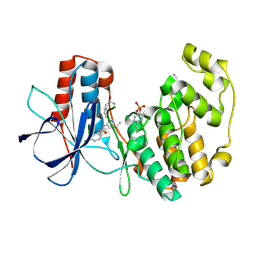 | | Crystal structure of phosphorylated P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNV
 
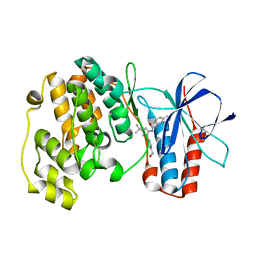 | | Crystal structure of P38 alpha in complex with DP437 | | Descriptor: | 1-{3-tert-butyl-1-[4-(hydroxymethyl)phenyl]-1H-pyrazol-5-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7JZU
 
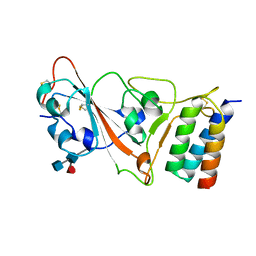 | |
7JZM
 
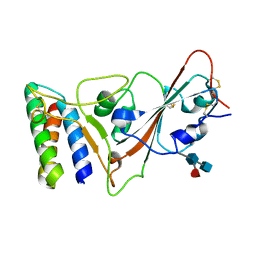 | |
8T5F
 
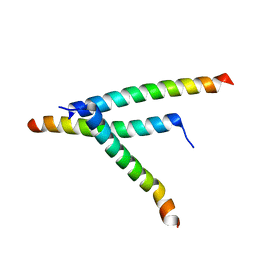 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Parathyroid hormone | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8T5E
 
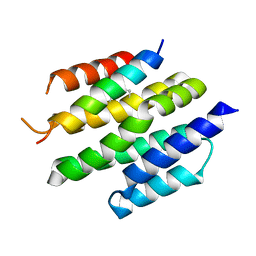 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Bcl-2-like protein 11, Bim_fulldiff | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
7UDK
 
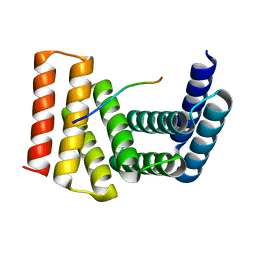 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UE2
 
 | |
7UDJ
 
 | | Crystal structure of designed helical repeat protein RPB_PEW3_R4 bound to PAWx4 peptide | | Descriptor: | 4xPAW peptide, De novo designed helical repeat protein RPB_PEW3_R4 | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
3UAM
 
 | |
3TSM
 
 | |
3U5W
 
 | |
7KNA
 
 | | Localized reconstruction of the H1 A/Michigan/45/2015 ectodomain displayed at the surface of I53_dn5 nanoparticle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,I53_dn5 | | Authors: | Acton, O.J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Quadrivalent influenza nanoparticle vaccines induce broad protection.
Nature, 592, 2021
|
|
3UJH
 
 | |
3U0I
 
 | |
