6NL6
 
 | | Crystal structure of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (T16F, T18A, V21E, T25L, K28Y, V29I, K31R, Q32H, Y33L, N35K, D36H, N37Q) | | Descriptor: | CHLORIDE ION, Immunoglobulin G-binding protein G, ZINC ION | | Authors: | Huxford, T, Stec, B, Maniaci, B. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
6NL8
 
 | | Crystal structure of de novo designed metal-controlled dimer of mutant B1 immunoglobulin-binding domain of Streptococcal Protein G (L12H, T16L, V29H, Y33H, N37L)-zinc | | Descriptor: | CHLORIDE ION, Immunoglobulin G-binding protein G, ZINC ION | | Authors: | Maniaci, B, Stec, B, Huxford, T. | | Deposit date: | 2019-01-08 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of High-Affinity Metal-Controlled Protein Dimers.
Biochemistry, 58, 2019
|
|
4ENL
 
 | |
5UDG
 
 | | Mutant E97Q crystal structure of Bacillus subtilis QueF with a disulfide Cys 55-99 | | Descriptor: | MAGNESIUM ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, TRIETHYLENE GLYCOL | | Authors: | Mohammad, A, Kiani, M.K, Iwata-Reuyl, D, Stec, B, Swairjo, M. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide.
Biomolecules, 7, 2017
|
|
3HDI
 
 | | Crystal structure of Bacillus halodurans metallo peptidase | | Descriptor: | COBALT (II) ION, Processing protease, SULFATE ION, ... | | Authors: | Aleshin, A, Gramatikova, S, Strongin, A.Y, Stec, B, Liddington, R.C, Smith, J.W. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal and solution structures of a prokaryotic M16B peptidase: an open and shut case.
Structure, 17, 2009
|
|
3L4C
 
 | | Structural basis of membrane-targeting by Dock180 | | Descriptor: | BETA-MERCAPTOETHANOL, Dedicator of cytokinesis protein 1 | | Authors: | Premkumar, L, Bobkov, A.A, Patel, M, Jaroszewski, L, Bankston, L.A, Stec, B, Vuori, K, Cote, J.-F, Liddington, R.C. | | Deposit date: | 2009-12-18 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of membrane targeting by the Dock180 family of Rho family guanine exchange factors (Rho-GEFs).
J.Biol.Chem., 285, 2010
|
|
3FKU
 
 | | Crystal structure of influenza hemagglutinin (H5) in complex with a broadly neutralizing antibody F10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Neutralizing antibody F10, ... | | Authors: | Hwang, W.C, Santelli, E, Stec, B, Wei, G, Cadwell, G, Bankston, L.A, Sui, J, Perez, S, Aird, D, Chen, L.M, Ali, M, Murakami, A, Yammanuru, A, Han, T, Cox, N, Donis, R.O, Liddington, R.C, Marasco, W.A. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3EZQ
 
 | | Crystal Structure of the Fas/FADD Death Domain Complex | | Descriptor: | Protein FADD, SODIUM ION, SULFATE ION, ... | | Authors: | Schwarzenbacher, R, Robinson, H, Stec, B, Riedl, S.J. | | Deposit date: | 2008-10-23 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Fas-FADD death domain complex structure unravels signalling by receptor clustering
Nature, 457, 2009
|
|
3T5O
 
 | | Crystal Structure of human Complement Component C6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, Complement component C6, ... | | Authors: | Aleshin, A.E, Stec, B, Bankston, L.A, DiScipio, R.G, Liddington, R.C. | | Deposit date: | 2011-07-27 | | Release date: | 2012-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.869 Å) | | Cite: | Structure of Complement C6 Suggests a Mechanism for Initiation and Unidirectional, Sequential Assembly of Membrane Attack Complex (MAC).
J.Biol.Chem., 287, 2012
|
|
3V16
 
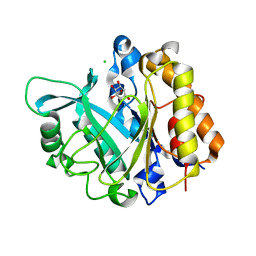 | | An intramolecular pi-cation latch in phosphatidylinositol-specific phospholipase C from S.aureus controls substrate access to the active site | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
3V18
 
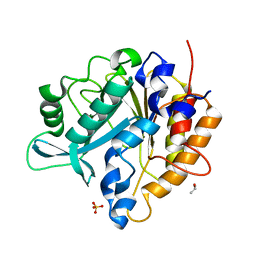 | | Structure of the Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ISOPROPYL ALCOHOL, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
3V1H
 
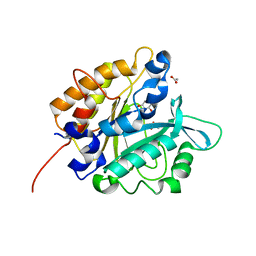 | | Structure of the H258Y mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Roberts, M.F. | | Deposit date: | 2011-12-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the S. aureus PI-Specific Phospholipase C Reveals Modulation of Active Site Access by a Titratable PI-Cation Latched Loop
Biochemistry, 51, 2012
|
|
4E0S
 
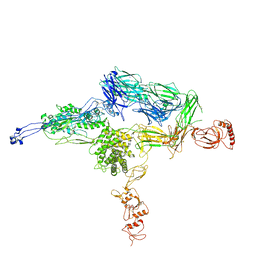 | | Crystal Structure of C5b-6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C5, ... | | Authors: | Aleshin, A.E, Stec, B, DiScipio, R, Liddington, R.C. | | Deposit date: | 2012-03-05 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.21 Å) | | Cite: | Crystal structure of c5b-6 suggests structural basis for priming assembly of the membrane attack complex.
J.Biol.Chem., 287, 2012
|
|
6N9A
 
 | |
2P3N
 
 | | Thermotoga maritima IMPase TM1415 | | Descriptor: | Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Stieglitz, K.A, Roberts, M.F, Li, W, Stec, B. | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J., 274, 2007
|
|
2P3V
 
 | | Thermotoga maritima IMPase TM1415 | | Descriptor: | Inositol-1-monophosphatase, S,R MESO-TARTARIC ACID | | Authors: | Stieglitz, K.A, Roberts, M.F, Li, W, Stec, B. | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the tetrameric inositol 1-phosphate phosphatase (TM1415) from the hyperthermophile, Thermotoga maritima.
Febs J., 274, 2007
|
|
2PLH
 
 | | STRUCTURE OF ALPHA-1-PUROTHIONIN AT ROOM TEMPERATURE AND 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2-BUTANOL, ACETATE ION, ALPHA-1-PUROTHIONIN, ... | | Authors: | Teeter, M.M, Stec, B, Rao, U. | | Deposit date: | 1993-07-09 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of purothionins reveals solute particles important for lattice formation and toxicity. Part 1: alpha1-purothionin revisited.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2QFL
 
 | | Structure of SuhB: Inositol monophosphatase and extragenic suppressor from E. coli | | Descriptor: | ACETATE ION, ETHYL ACETATE, Inositol-1-monophosphatase | | Authors: | Wang, Y, Stieglitz, K.A, Bubunenko, M, Court, D, Stec, B, Roberts, M.F. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the R184A mutant of the inositol monophosphatase encoded by suhB and implications for its functional interactions in Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
1EW9
 
 | | ALKALINE PHOSPHATASE (E.C. 3.1.3.1) COMPLEX WITH MERCAPTOMETHYL PHOSPHONATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, MERCAPTOMETHYL PHOSPHONATE, ... | | Authors: | Holtz, K.M, Stec, B, Meyers, J.K, Antonelli, S.M, Widlanski, T.S, Kantrowitz, E.R. | | Deposit date: | 2000-04-24 | | Release date: | 2002-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alternate modes of binding in two crystal structures of alkaline phosphatase-inhibitor complexes.
Protein Sci., 9, 2000
|
|
1EW8
 
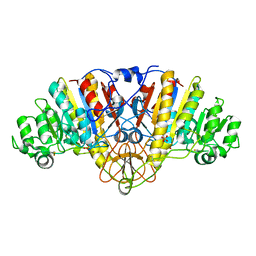 | | ALKALINE PHOSPHATASE (E.C. 3.1.3.1) COMPLEX WITH PHOSPHONOACETIC ACID | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Holtz, K.M, Stec, B, Myers, J.K, Antonelli, S.M, Widlanski, T.S, Kantrowitz, E.R. | | Deposit date: | 2000-04-24 | | Release date: | 2002-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alternate modes of binding in two crystal structures of alkaline phosphatase-inhibitor complexes.
Protein Sci., 9, 2000
|
|
2GHW
 
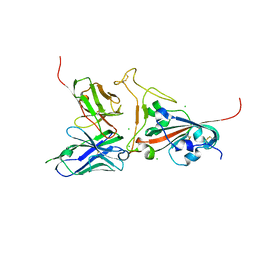 | | Crystal structure of SARS spike protein receptor binding domain in complex with a neutralizing antibody, 80R | | Descriptor: | CHLORIDE ION, Spike glycoprotein, anti-sars scFv antibody, ... | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2GHV
 
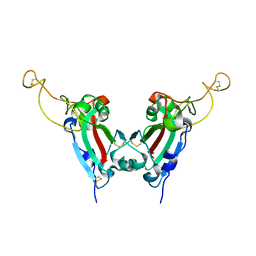 | | Crystal structure of SARS spike protein receptor binding domain | | Descriptor: | Spike glycoprotein | | Authors: | Hwang, W.C, Lin, Y, Santelli, E, Sui, J, Jaroszewski, L, Stec, B, Farzan, M, Marasco, W.A, Liddington, R.C. | | Deposit date: | 2006-03-27 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R.
J.Biol.Chem., 281, 2006
|
|
2I39
 
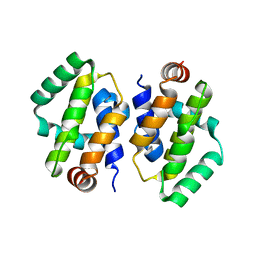 | | Crystal structure of Vaccinia virus N1L protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein N1 | | Authors: | Aoyagi, M, Aleshin, A.E, Stec, B, Liddington, R.C. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein.
Protein Sci., 16, 2007
|
|
1LBY
 
 | | Crystal Structure of a complex (P32 crystal form) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus with 3 Manganese ions, Fructose-6-Phosphate, and Phosphate ion | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
1LBV
 
 | | Crystal Structure of apo-form (P21) of dual activity FBPase/IMPase (AF2372) from Archaeoglobus fulgidus | | Descriptor: | fructose 1,6-bisphosphatase/inositol monophosphatase | | Authors: | Stieglitz, K.A, Johnson, K.A, Yang, H, Roberts, M.F, Seaton, B.A, Head, J.F, Stec, B. | | Deposit date: | 2002-04-04 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a dual activity IMPase/FBPase (AF2372) from Archaeoglobus fulgidus. The story of a mobile loop.
J.Biol.Chem., 277, 2002
|
|
