4IMQ
 
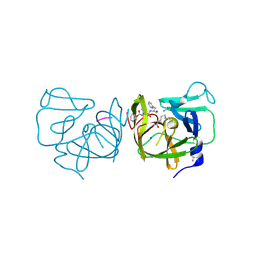 | | Structural Basis of Substrate Specificity and Protease Inhibition in Norwalk Virus | | Descriptor: | 3C-like protease, PEPTIDE INHIBITOR, syc8, ... | | Authors: | Prasad, B.V.V, Muhaxhiri, Z, Deng, L, Shanker, S, Sankaran, B, Estes, M.K, Palzkill, T, Song, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of substrate specificity and protease inhibition in norwalk virus.
J.Virol., 87, 2013
|
|
6MO1
 
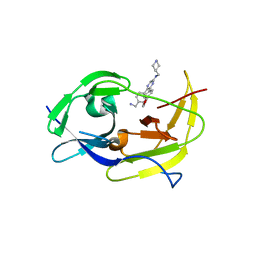 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 5-[4-(aminomethyl)phenyl]-6-[4-(furan-3-yl)phenyl]-N-[(piperidin-4-yl)methyl]pyrazin-2-amine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Hua, Y, Nie, S, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
3ANN
 
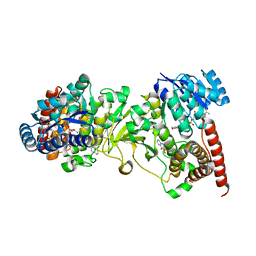 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with quinolin-2-ylmethylphosphonic acid | | Descriptor: | (quinolin-2-ylmethyl)phosphonic acid, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Endo, K, Kato, M, Deng, L, Song, Y, Yajima, S. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase/Lipophilic Phosphonate Complexes
ACS Med Chem Lett, 2, 2011
|
|
3ANM
 
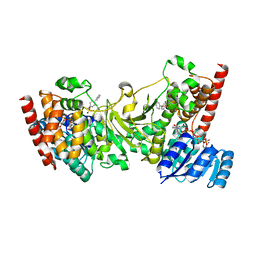 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with 5-phenylpyridin-2-ylmethylphosphonic acid | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [(5-phenylpyridin-2-yl)methyl]phosphonic acid | | Authors: | Endo, K, Kato, M, Deng, L, Song, Y, Yajima, S. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase/Lipophilic Phosphonate Complexes
ACS Med Chem Lett, 2, 2011
|
|
6MO0
 
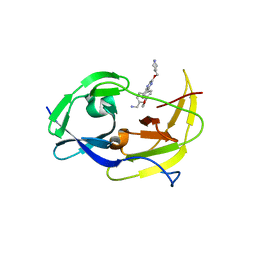 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{3-[4-(furan-3-yl)phenyl]-5-[(piperidin-4-yl)methoxy]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J. Am. Chem. Soc., 141, 2019
|
|
3ANL
 
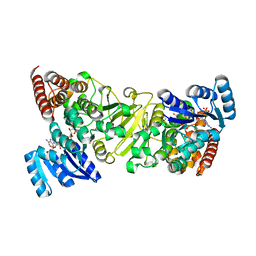 | | Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) complexed with pyridin-2-ylmethylphosphonic acid | | Descriptor: | (pyridin-2-ylmethyl)phosphonic acid, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Endo, K, Kato, M, Deng, L, Song, Y, Yajima, S. | | Deposit date: | 2010-09-03 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase/Lipophilic Phosphonate Complexes
ACS Med Chem Lett, 2, 2011
|
|
6MO2
 
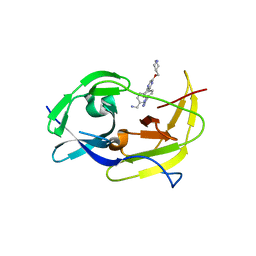 | | Structure of dengue virus protease with an allosteric Inhibitor that blocks replication | | Descriptor: | 1-(4-{5-[(piperidin-4-yl)methoxy]-3-[4-(1H-pyrazol-4-yl)phenyl]pyrazin-2-yl}phenyl)methanamine, FLAVIVIRUS_NS2B/Peptidase S7 | | Authors: | Lin, Y.-L, Nie, S, Hua, Y, Wu, J, Wu, F, Huo, T, Yao, Y, Song, Y. | | Deposit date: | 2018-10-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease.
J.Am.Chem.Soc., 141, 2019
|
|
4S0J
 
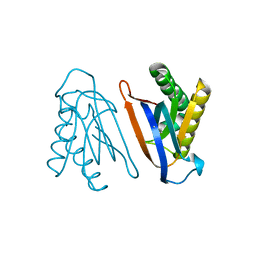 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123V mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S03
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, Y79I, and F123A mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S0L
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, Y79V, and F123V mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S02
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, F42W, Y79A, and F123Y mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S0K
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79V, and 123A mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S0I
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123A mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
2LP1
 
 | | The solution NMR structure of the transmembrane C-terminal domain of the amyloid precursor protein (C99) | | Descriptor: | C99 | | Authors: | Barrett, P.J, Song, Y, Van Horn, W.D, Hustedt, E.J, Schafer, J.M, Hadziselimovic, A, Beel, A.J, Sanders, C.R. | | Deposit date: | 2012-01-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol.
Science, 336, 2012
|
|
2MV0
 
 | | Solution NMR Structure of Maltose-binding protein from Escherichia coli, Northeast Structural Genomics Consortium (NESG) Target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Rossi, P, Lange, O.F, Sgourakis, N.G, Song, Y, Lee, H, Aramini, J.M, Ertekin, A, Xiao, R, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2NDJ
 
 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
5HS5
 
 | |
5J08
 
 | |
5J2Y
 
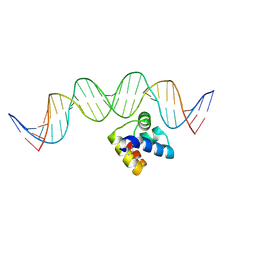 | | Molecular insight into the regulatory mechanism of the quorum-sensing repressor RsaL in Pseudomonas aeruginosa | | Descriptor: | DNA (26-MER), Regulatory protein | | Authors: | Zhao, J, Gan, J, Zhang, J, Kang, H, Kong, W, Zhu, M, Li, F, Song, Y, Qin, J, Liang, H. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing.
Nucleic Acids Res., 45, 2017
|
|
5CB3
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CMW
 
 | | Crystal structure of yeast Ent5 N-terminal domain-soaked in KI | | Descriptor: | Epsin-5, GLYCEROL, IODIDE ION | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5CN2
 
 | |
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
5CN1
 
 | |
5CMY
 
 | | Crystal structure of yeast Ent5 N-terminal domain-native | | Descriptor: | Epsin-5, GLYCEROL | | Authors: | Zhang, F, Song, Y, Li, X, Teng, M.K. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and functional insight into the N-terminal domain of the clathrin adaptor Ent5 from Saccharomyces cerevisiae
Biochem.Biophys.Res.Commun., 477, 2016
|
|
